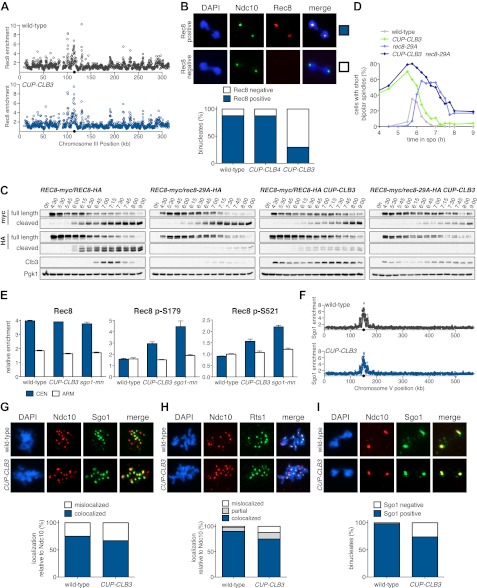
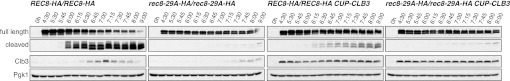
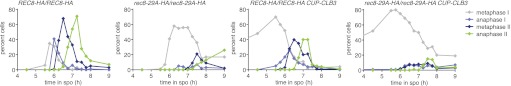
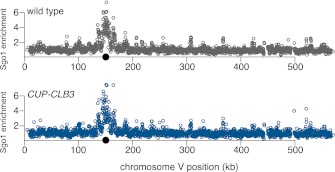
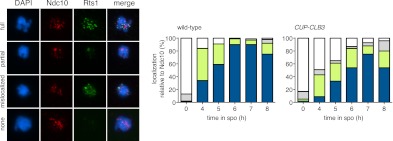
Figure 4. CLB3 misexpression disrupts protection of centromeric cohesin.
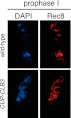
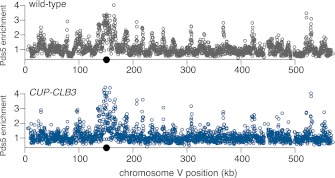
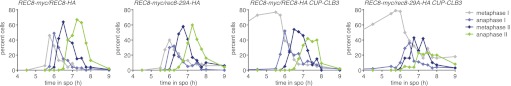
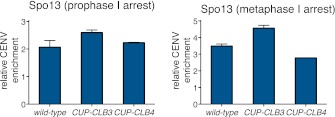
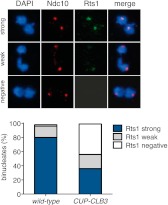
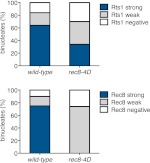
Cyclin expression was induced after 2 hr 15 min (C) and (D), 2 hr 30 min (A), (B), (E), (F) and (H) or 3 hr (G) and (I) of sporulation. (A) Chromosomal association of Rec8-13myc was monitored by ChIP-chip in wild-type (A28716) and CUP-CLB3 (A28718) during prophase I arrest. Centromere position is identified by a black circle. (B) Centromeric Rec8 localization was monitored in spread nuclei from wild-type (A28684), CUP-CLB3 (A28685) and CUP-CLB4 (A28686) cells carrying REC8-3HA (red) and NDC10-13myc (green) (n > 40). The fraction of spread nuclei that were Rec8 positive or negative was compared to wild-type using a chi-square test (df 1): CUP-CLB4, χ2 = 0.001323, p=0.9710; CUP-CLB3, χ2 = 32.79, p<0.0001. (C) Rec8 cleavage monitored by Western blot after release from an NDT80 block (4 hr 30 min) in wild-type and CUP-CLB3 carrying both a myc-tagged REC8 allele as well as either HA-tagged REC8 or rec8-29A allele (left to right: A29957, A29959, A29961, A29963). (D) Percentage of cells with short bipolar spindles was determined at indicated times in wild-type (A22804), CUP-CLB3 (A29965), rec8-29A (A22803) and CUP-CLB3 rec8-29A (A29967) after release from an NDT80 block (4 hr 30 min) (n = 100 per time point). (E) ChIP analysis for total Rec8, p-S179 Rec8 or p-S521 Rec8 from metaphase I-arrested (cdc20-mn) wild-type (A28681), CUP-CLB3 (A28682) and Sgo1-depleted (sgo1-mn; A29994) cells. Relative occupancy at a chromosome arm site (c194) or at a centromeric site (CENV) was determined relative to a low binding region (c281). Error bars represent range (n = 2). (F) Chromosomal association of Sgo1-3V5 was monitored by ChIP-chip in wild-type (A29795) and CUP-CLB3 (A29799) cells during prophase I-arrest. Centromere position is identified by a black circle. (G), (H) Localization of Sgo1-9myc (G, green) or Rts1-13myc (H, green) relative to Ndc10-6HA (red) determined by nuclear spreads in (G) wild-type (A22868) and CUP-CLB3 (A22870) or (H) wild-type (A28329) and CUP-CLB3 (A28330) during prophase I (n > 40). For (G), the fraction of spread nuclei that display colocalized or mislocalized Sgo1 relative to Ndc10 was compared between wild-type and CUP-CLB3 using a chi-square test (df 1) χ2 = 1.554, p=0.2125. For (H), the fraction of spread nuclei that display colocalized, partial or mislocalized Rts1 relative to Ndc10 was compared between wild-type and CUP-CLB3 using a chi-square test (df 2) χ2 = 3.712, p=0.1563. (I) Localization of Sgo1-9myc (green) in binucleates relative to Ndc10-6HA (red) determined by nuclear spreads from wild-type (A22868) and CUP-CLB3 (A22870) (n > 40). The fraction of spread nuclei that were Sgo1 positive or negative was compared between wild-type and CUP-CLB3 using a chi-square test (df 1) χ2 = 23.92, p<0.0001.
Figure 4—figure supplement 1. Chromosomal association of Rec8 in CUP-CLB3 cells.