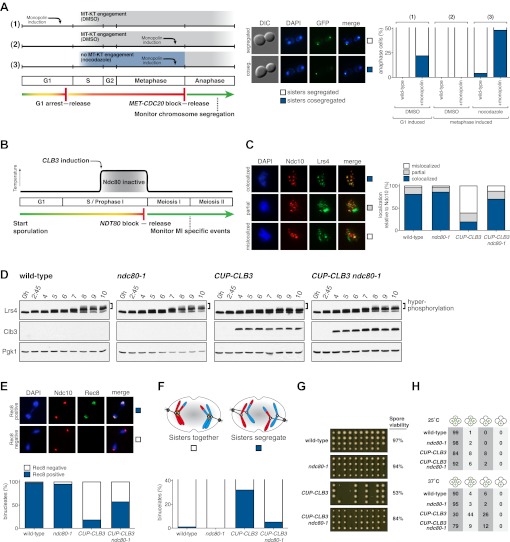
Figure 5. Transient disruption of microtubule–kinetochore interactions suppresses the chromosome segregation defects in CUP-CLB3 cells.
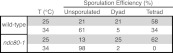
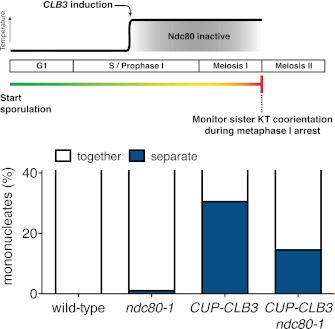
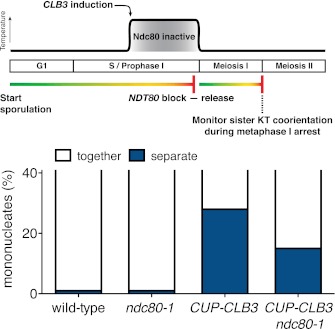
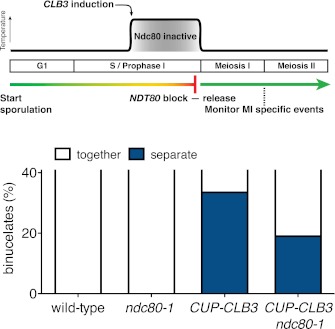
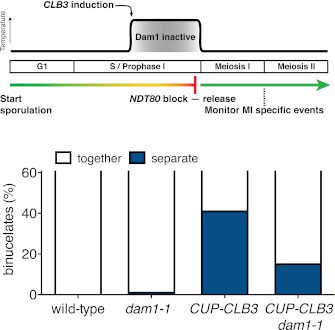
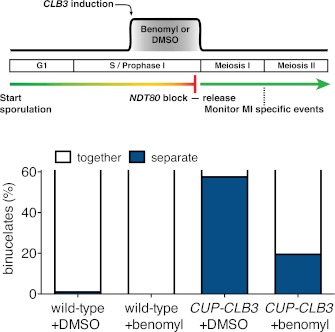
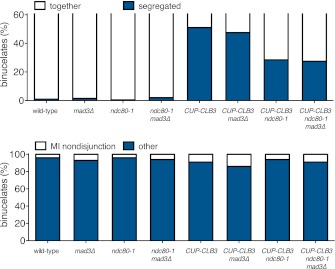
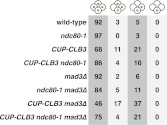
(A) Wild-type (A10684) and GAL-CDC5 GAL-MAM1 (A26546) cells, carrying a MET-CDC20 allele and CENIV-GFP dots, were monitored for chromosome segregation in anaphase (see ‘Materials and methods’ for details). MT = microtubule, KT = kinetochore, (n = 100). The fraction of anaphase cells that segregate or cosegregate sister chromatids was compared between GAL-CDC5 GAL-MAM1 condition (2) and GAL-CDC5 GAL-MAM1 condition (3) using a chi-square test (df 1) χ2 = 59.71, p<0.0001. (B) Schematic description of the experimental regime used for (C) through (H) see ‘Materials and methods’ for details. (C) Localization of Lrs4-13myc (green) in mononucleates relative to Ndc10-6HA (red) determined by nuclear spreads (n > 40) and (D) phosphorylation of Lrs4-13myc determined by gel mobility shift in wild-type (A29612), ndc80-1 (A29614), CUP-CLB3 (A29616) and CUP-CLB3 ndc80-1 (A29618). For (C), using a chi-square test (df 2) the fraction of spread nuclei that display colocalized, partial or mislocalized Lrs4 with respect to Ndc10 was compared between wild-type and ndc80-1 χ2 = 0.9668, p=0.6167 and between CUP-CLB3 and CUP-CLB3 ndc80-1 χ2 = 56.34, p<0.0001. (E) Localization of Rec8-13myc (green) in binucleates relative to Ndc10-6HA (red) determined by nuclear spreads in wild-type (A28716), ndc80-1 (A28720), CUP-CLB3 (A28718) and CUP-CLB3 ndc80-1 (A28722) (n > 40). Using a chi-square test (df 1) the fraction of spread nuclei that were Rec8 positive or negative was compared between wild-type and ndc80-1 χ2 = 1.185, p=0.2764 and between CUP-CLB3 and CUP-CLB3 ndc80-1 χ2 = 23.96, p<0.0001. (F) Segregation of sister chromatids using heterozygous CENV-GFP dots quantified in binucleates (n = 100) and (G) spore viability from wild-type (A22678), ndc80-1 (A28621), CUP-CLB3 (A22702) and CUP-CLB3 ndc80-1 (A28623) (n = 40 tetrads for wild-type and ndc80-1, n > 60 tetrads for CUP-CLB3 and CUP-CLB3 ndc80-1) (nonpermissive temperature >36°C). Using a chi-square test (df 1) the fraction of binucleates with a reductional or equational division was compared between CUP-CLB3 and CUP-CLB3 ndc80-1 χ2 = 24.18, p<0.0001. (G) Segregation of chromosome V using homozygous CENV-GFP dots quantified in tetranucleates from wild-type (A22688), ndc80-1 (A28625), CUP-CLB3 (A22708) and CUP-CLB3 ndc80-1 (A28627). Top panel: cells kept at 25°C for the duration of the experiment. Bottom panel: Cells treated as in (B) but monitored after meiosis II (n = 100).