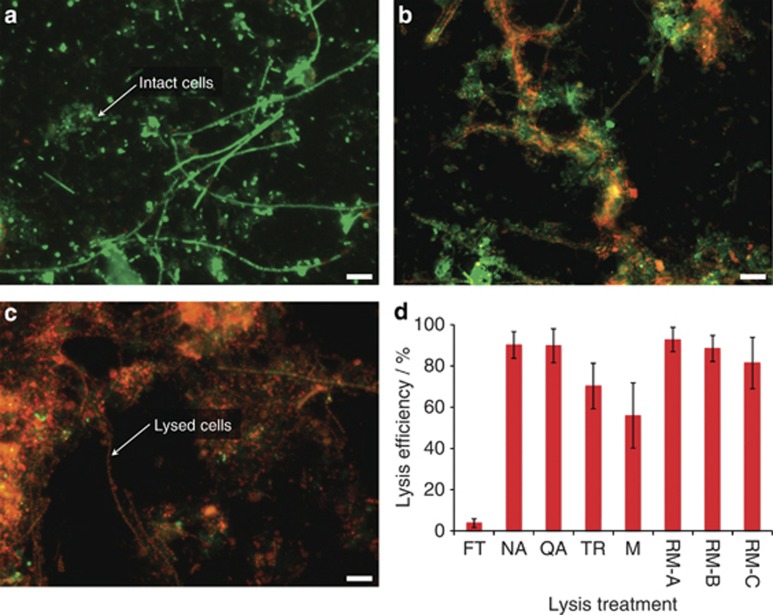
Figure 3.
Efficiencies of different cell lysis methods. (a–c) Representative epifluorescence micrographs of microbial cells from a representative LAO-enriched microbial community sample stained with the Live/Dead stain (intact cells highlighted in green, disrupted cells with a compromised cell membrane in red). Scale bar is equivalent to 10 μm. (a) Sample having undergone a single freeze-thaw cycle. (b) Sample having undergone polar and non-polar metabolite extractions. (c) Sample having undergone the additional mechanical and chemical lysis step using the NA kit's modified lysis buffer. (d) Bar chart highlighting the percentages of cells disrupted by the different treatments (n=10; error bars represent s.d.). x-axis legend: FT, sample having undergone a single freeze-thaw cycle, reflecting panel a; NA, sample having been subjected to cell lysis in the modified NA lysis buffer, reflecting panel c; QA, QA lysis buffer; TR, TR-based lysis; M, sample after polar and non-polar metabolite extractions reflecting panel b; RM-A, sample having been subjected to the dedicated DNA extraction reference method; RM-B, dedicated RNA extraction method; RM-C, dedicated protein extraction method.