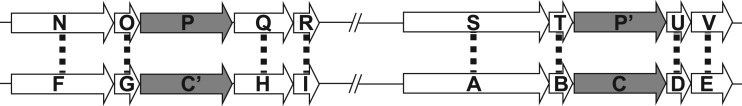
Figure 1.
An example of how CGN can be used to cluster paralogous genes into orthologous pairs. Open arrows indicate proteins, with dotted lines illustrating best BLAST matches. Gray arrows indicate paralogous proteins with multiple high-identity BLAST matches. The genes on top are from one genome and those on the bottom from a second genome. The slashes indicated that there are genes in between not drawn.