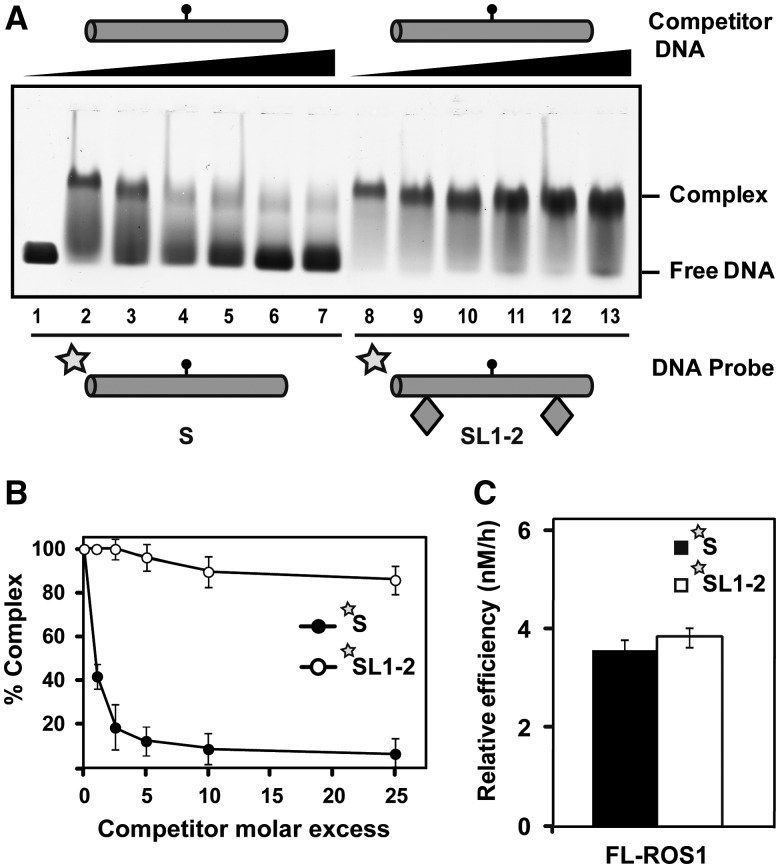
Figure 5.
ROS1 linear diffusion along DNA does not facilitate processing of T:G mispairs. (A) Gel shift assay showing dissociation of FL-ROS1 (120 nM) from fluorescein-labelled substrates S (lanes 2–7, 100 nM) and SL1-2 (lanes 8–13, 100 nM) containing a T:G mispair on addition of increasing amounts (0, 0.1, 0.25, 0.5, 1.0 and 2.5 μM) of a mismatched unlabelled S competitor. After non-denaturing gel electrophoresis, the gel was scanned to detect fluorescein-labelled DNA. Protein-DNA complexes were identified by their retarded mobility compared with that of free DNA, as indicated. (B) Graph shows the percentage of remaining complex FL-ROS1-DNA versus competitor molar excess ratios. Values are mean ± SE from two independent experiments. (C) FL-ROS1 (20 nM) was incubated at 30°C with fluorescein-labelled substrates S and SL1-2 (20 nM) containing a single T:G mispair. Reaction products were separated in a 12% denaturing polyacrylamide gel and quantified by fluorescence scanning. Values are mean ± SE of the relative processing efficiencies (Erel) from two independent experiments, adjusted with the unbiased estimator described in (36).