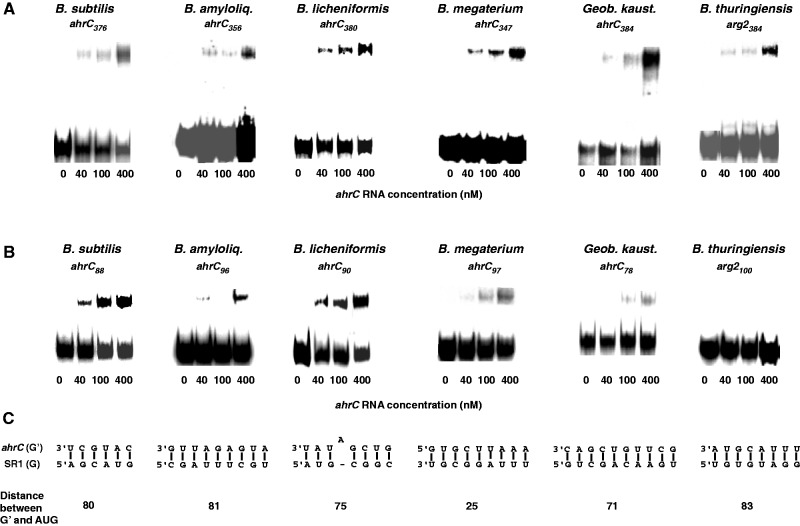
Figure 3.
Binding assays of SR1/ahrC RNA pairs from different Bacillus species. Binding experiments were performed as described in Materials and Methods section. Autoradiograms of gel-shift assays are shown. The concentration of unlabelled ahrC species is indicated in nM. (A) Binding assays with homologous SR1/ahrC pairs from B. subtilis, B. amyloliquefaciens, B. licheniformis, B. megaterium, Geob. kaustophilus and B. thuringiensis III. SR1 species from these Bacilli were 5′ end-labelled with [γ32P]-ATP and used at a final concentration of 4 nM (at least 10-fold lower concentration than ahrC RNA) in all experiments. ahrC-RNA species of the indicated length comprising the central part of ahrC with all complementary regions (Supplementary Figures S7 and S8) were used. (B) Binding assays with homologous SR1/ahrC pairs as above. ahrC RNAs containing only region G′ were paired with the SR1 homologues comprising all complementary regions. (C) Regions that can base pair in the experiment shown in (B). Below, the distance between the 5′ end of region G′ and the 3′ end of the corresponding ahrC start codons is shown for each ahrC RNA/SR1 pair.