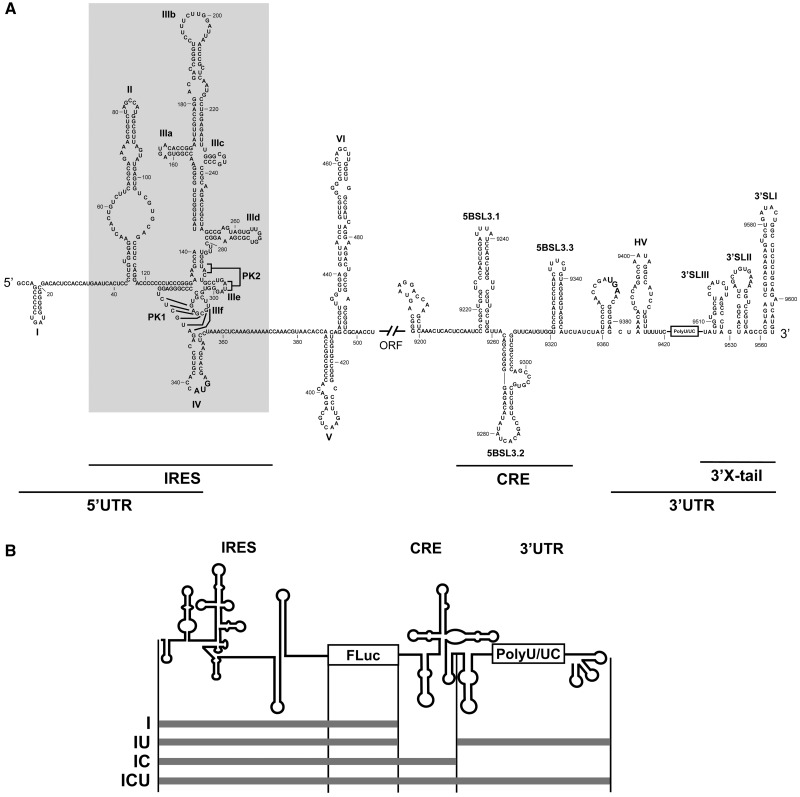
Figure 1.
Secondary structure of the 5′- and 3′-ends of the HCV genome. (A) Sequence and secondary structure of the 5′- and 3′-regions of the HCV genome. The 5′-UTR plus domains V and VI located inside the HCV coding sequence are included. The minimum region for IRES activity is boxed. The 3′-end of the viral genomic RNA is organized into two structural elements: the CRE region and the 3′ X-tail, separated by a hypervariable sequence (HV) and a polyU/UC stretch (also considered inside the 3′-UTR). Numbers refer to the nucleotide positions of the HCV Con1 isolate (GenBank accession number AJ238799). Start and stop translation codons placed at positions 342 and 9371, respectively, are shown in bold. Pseudoknot elements are marked as PK1 and PK2. (B) Diagram of the HCV transcripts harboring the IRES–FLuc fusion attached to different 3′ viral genomic domains.