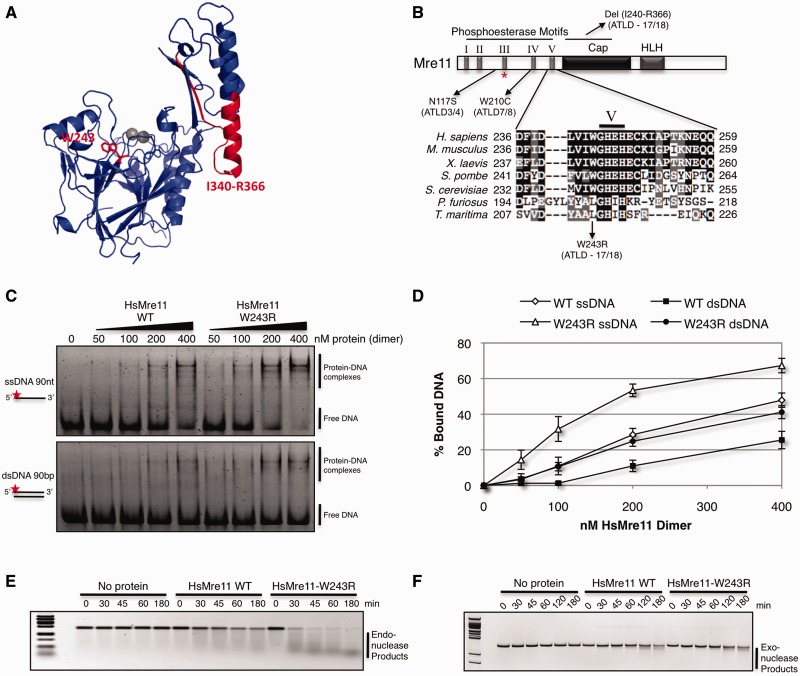
Figure 1.
Human Mre11-W243R is proficient in nuclease activities. (A) Structure of human Mre11 monomer (PDB: 3T1I) with residues deleted or mutated in ATLD17/18 highlighted in red. (B) Mre11 schematic domain architecture and sequence alignment showing location of ATLD mutations and local alignment at conserved hydrophobic site W243. Mre11-W243 is adjacent to conserved nuclease motif V. HLH motif binds base of Rad50 coiled coil, forming the main interaction interface. Red asterisk (*) denotes HsMre11 histidine 129 (SpMre11-H134, PfMre11-H85) mutated in nuclease dead alleles. (C) EMSA of increasing amounts of purified HsMre11 incubated with ssDNA or dsDNA substrate labeled with Alexa Fluor 488 separated by native PAGE. (D) Quantification of free DNA from representative gel in panel D expressed as percentage of bound DNA. Error bars represent standard error of the mean of four independent experiments. (E) Endonuclease activity of HsMre11 on circular ssDNA plasmid substrate, ϕX174. Reactions were carried out at 37°C for the indicated time points. (F) Exonuclease activity of HsMre11 on linearized double-stranded plasmid DNA substrate pBluescript. Reactions were carried out at 37°C for the indicated time points.