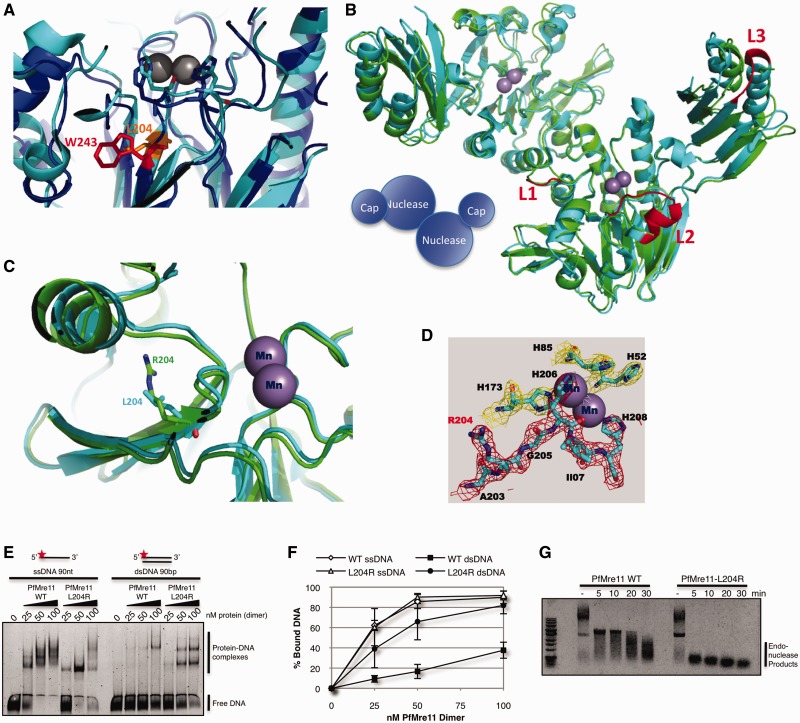
Figure 2.
X-ray crystal structure of P. furiosus Mre11-L204R. (A) Detailed view of nuclease core of HsMre11 (PDB: 3T1I) (blue) superimposed with PfMre11 (PDB: 1II7) (cyan) with Trp243 (human, red) and Leu204 (Pyrococcus, orange) highlighted. (B) Structure of PfMre11-L204R dimer (PDB: 4HD0) (green) superimposed on wild-type PfMre11 dimer (PDB: 1II7) (cyan). Mn+2 ions in active site are shown as purple spheres. Unfolded (disordered) loops in PfMre11-L204R are highlighted in red on the wild-type PfMre11 structure (C) Details of active site of wild-type PfMre11 and PfMre11-L204R. (D) 2Fo-Fc electron density map showing the PfMre11-L204R active site. (E) EMSA of increasing amounts purified PfMre11 incubated with single- or double- stranded DNA labeled with Alexa Fluor 488 (red star) separated by native PAGE. (F) Quantification of free DNA from representative gel in panel A expressed as percentage of bound DNA. Error bars represent standard error of the mean of four independent experiments. (G) Endonuclease activity of PfMre11 on circular ssDNA plasmid substrate, ϕX174. Reactions were carried out at 50°C for the indicated time points. Lanes marked—are DNA alone.