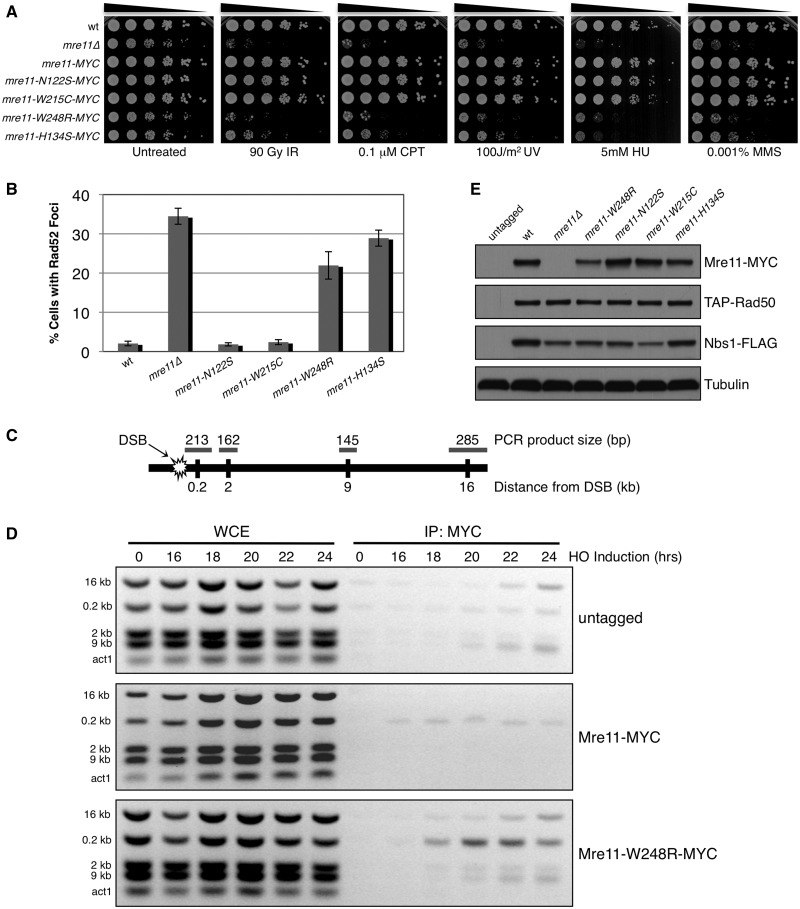
Figure 3.
Analysis of Mre11 ATLD site mutant in S. pombe. (A) 5-fold serial dilutions of Mre11 ATLD and nuclease dead (H134S) mutants were spotted and treated with indicated dose of genotoxic agents. The MYC-tagged Mre11 behaved similarly to the wild-type strain, indicating that the fusion does not alter protein function. IR = ionizing radiation, CPT = camptothecin, UV = ultraviolet radiation, HU = hydroxyurea, MMS = methyl methanesulfonate. (B) Percentage of cells with one or more spontaneously occurring Rad52-RFP focus. At least 1200 cells were analysed for each strain with error bars representing standard deviation from the mean of three experiments. (C) Schematic of chromosome I containing HO break site used in ChIP experiments indicating relative distances and expected sizes assayed by multiplex PCR. (D) ChIP of untagged, Mre11-MYC and Mre11-W248R-MYC samples collected at indicated time points after removal of thiamine from the media, leading to HO endonuclease induction. Small increases in ChIP signals are present around the HO site in the untagged controls (E) Expression levels of endogenous MRN subunits in Mre11 mutant backgrounds. Tubulin is a loading control.