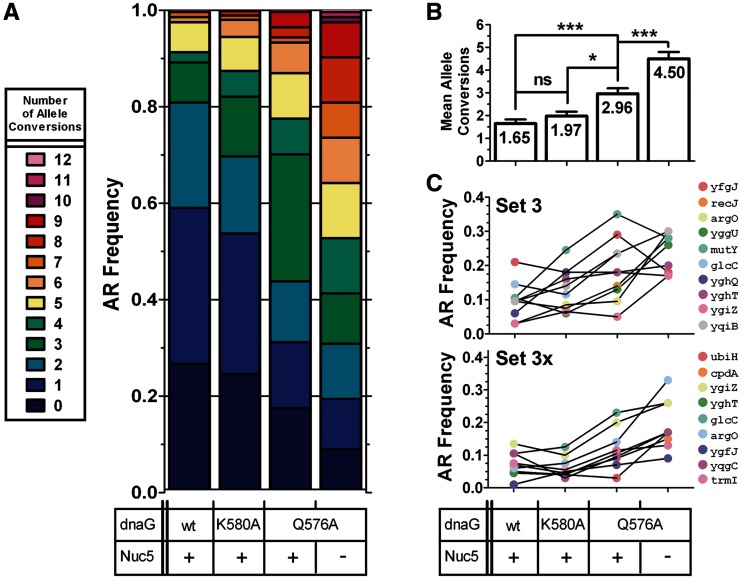
Figure 5.
Testing DnaG variants with a 20-plex CoS-MAGE oligo set. EcNR2, EcNR2.dnaG.K580A, EcNR2.dnaG.Q576A and Nuc5-.dnaG.Q576A were tested for their performance in CoS-MAGE using an expanded set of 20 oligos (Sets 3+3X). Genotypes of recombinant clones were assayed by mascPCR after one cycle of CoS-MAGE (ygfT could not be assayed by mascPCR). (A) AR frequency distributions. (B) Mean number of alleles converted ± SEM, with p-values indicated above the bars. Statistical significance is denoted using a star system where * denotes P < 0.003, ** denotes P < 0.001 and *** denotes P < 0.0001. (C) Mean individual AR frequencies. As seen with the smaller oligo sets, the dnaG variants reduce the number of clones with zero conversions and increase the average number of conversions per clone. Nuc5-.dnaG.Q576A strongly outperforms all other strains, with a mean of 4.50 alleles converted and fewer than 10% of clones having zero conversions. Notably, Nuc5-.dnaG.Q576A has strongly improved performance with Sets 3 + 3X compared with Set 3, whereas EcNR2.dnaG.Q576A does not. EcNR2, n = 96; EcNR2.dnaG.K580A, n = 113; EcNR2.dnaG.Q576A, n = 95; Nuc5-.dnaG.Q576A, n = 96.