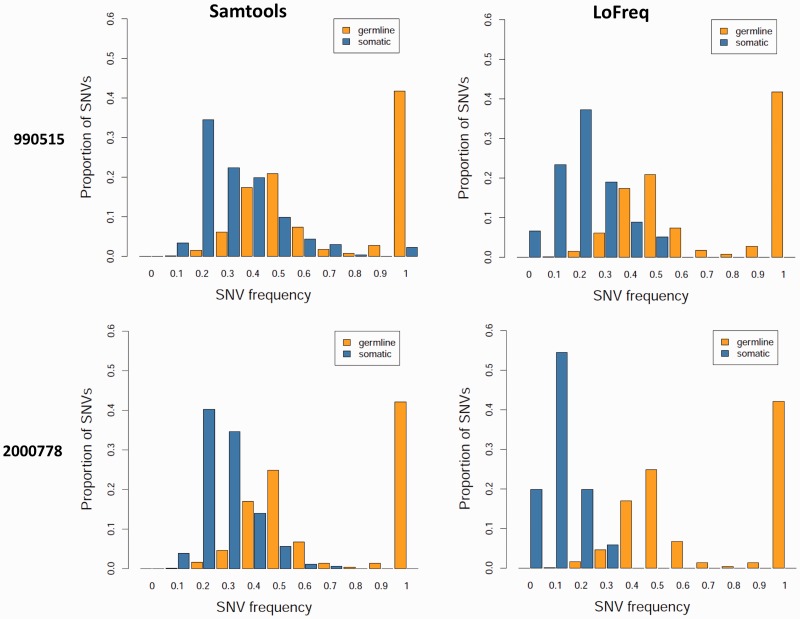
Figure 2.
SNV calling in the presence of tumor sample heterogeneity. Germline and somatic variant frequencies for paired tumor-normal exome sequencing datasets from a custom samtools-based pipeline (32) are compared here with those from LoFreq (see ‘Materials and Methods’ section). As shown, while germline variants are consistently distributed around 50% (as expected for heterozygous variants), somatic variants are shifted to lower frequencies, likely due to contamination in the tumor sample from normal stromal tissue. Note that while samtools-based somatic calls appear ‘clipped’ at lower frequencies, LoFreq calls are symmetrically distributed as expected.