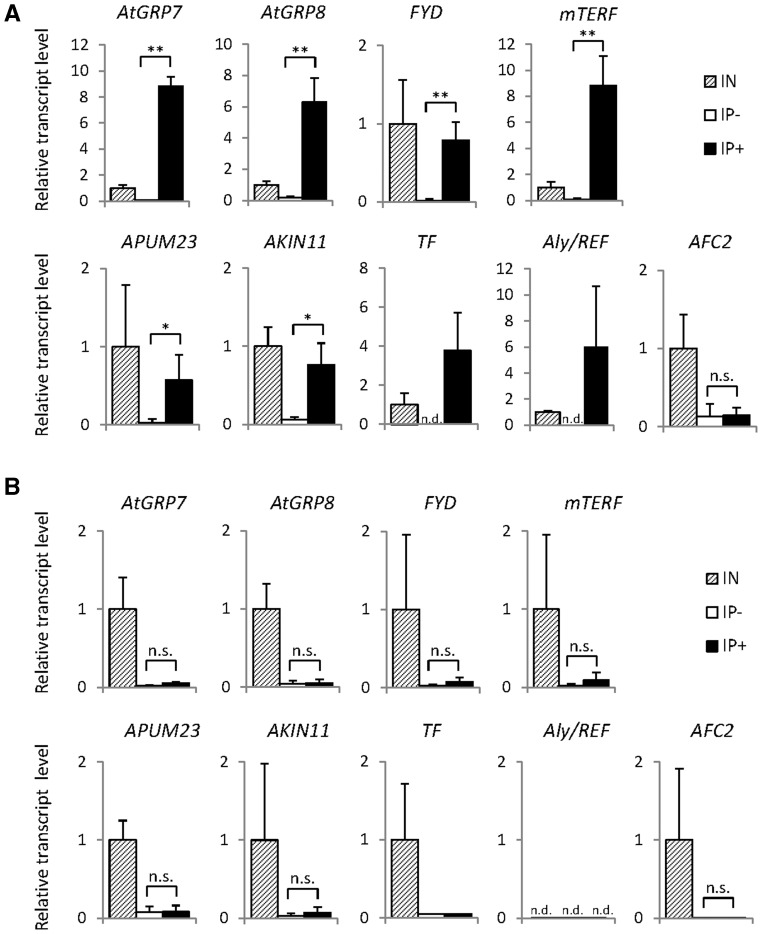
Figure 3.
In vivo interaction of AtGRP7-GFP with candidate target transcripts. RIP was performed on plants expressing the AtGRP7-GFP fusion protein under control of the AtGRP7 promoter including 5′- and 3′-UTR and intron in atgrp7-1 (A) and transgenic plants expressing GFP under control of the AtGRP7 promoter including 5′- and 3′-UTR (B). The levels of transcripts co-precipitated in the GFP-Trap® bead precipitate (IP+), the RFP-Trap® bead mock precipitate (IP−) and in the input fractions, respectively, were determined by qRT-PCR in duplicates for At2g21660 (AtGRP7) which served as positive control, At4g39260 (AtGRP8, #90), At3g12570 (FYD, #288), At2g3600 (mTERF, #75), At1g72320 (APUM23, #12), At3g29160 (AKIN11, #343), At5g05550 [transcription factor (TF), #181], At4g24740 (AFC2, #226 and #227) and At5g59950 (Aly/REF-like RNA binding/export factor, #327). Transcript levels were normalized to PP2A and expressed relative to the input. Means ± SD are presented based on three biological and significance was tested using Student’s t-test (**P < 0.005, *P < 0.05). n.d., not detectable; n.s., not significant. In IP− from GFP plants, the transcripts #181 and #327 were not detectable and thus no statistical test was applied.