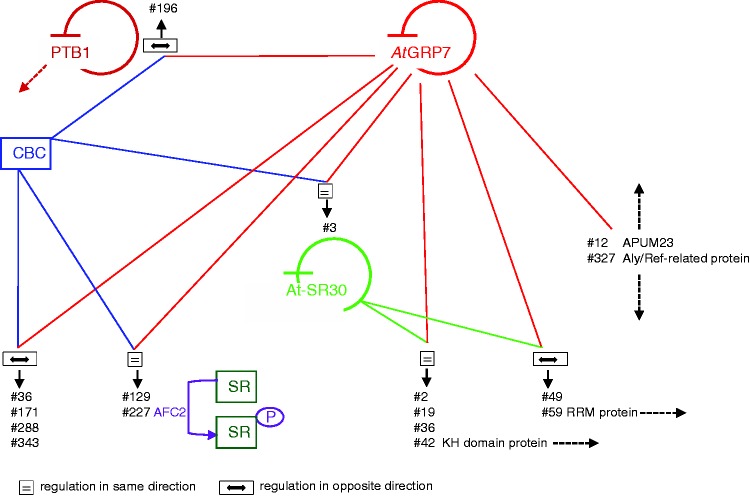
Figure 5.
Conceptual model depicting common targets of known splicing regulators in Arabidopsis. The impact of AtGRP7, the cap binding complex (CBC) and At-SR30 on selected splicing events, analyzed by the RT-PCR panel, is displayed (33,34). The numbers of the primer pairs for detection of the AS events are indicated. Regulation of the ratio of splice isoforms in the same direction is indicated by ‘=’; regulation in opposite direction is indicated by a line with two arrowheads. For RNA-binding proteins that are influenced by AtGRP7, the name is indicated and dotted arrows indicate a presumed post-transcriptional regulation of yet unknown targets of these proteins. The negative autoregulation of AtGRP7, PTB1 and At-SR30 is depicted (19,31,57). ‘P’ denotes phosphorylation of SR proteins by the LAMMER kinase AFC2 (68). For clarity, the effects of AtGRP8 and At-RS2Z33 on AS events are omitted (see text for details).