Figure 1.
Comparison of Targets of Wild-Type and Mutant Rrp44
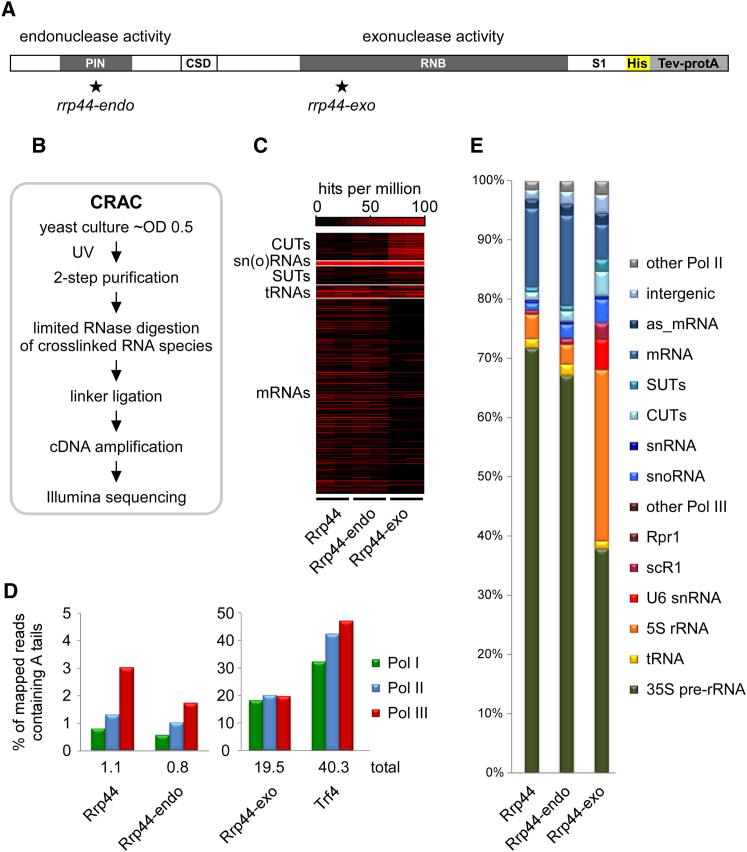
(A) Domain structure of S. cerevisiae Rrp44, including a C-terminal His-TEV protease-protein A (HTP) tag for purification. Point mutations inactivating the endonuclease (rrp44-endo) or exonuclease (rrp44-exo) activity of Rrp44 are indicated.
(B) Outline of the CRAC crosslinking technique.
(C–E) Illumina high-throughput sequencing of cDNA libraries generated from crosslinked RNAs recovered with purified wild-type Rrp44 and the Rrp44-endo and Rrp44-exo mutants, as well as the exosome cofactor Trf4. Here, and in all other illustrations, sequencing data of individual biological replicate experiments was mapped to the yeast genome using Novoalign and normalized to hits per million mapped sequences (hpm).
(C) Heat maps for main substrate groups. Numbers of reads mapped to individual RNAs are shown in shades of red.
(D) Frequencies of non-templated terminal oligo(A) sequence reads in data sets for wild-type Rrp44 and catalytic mutants, and the exosome cofactor Trf4. Data sets are filtered either for total reads, or for Pol I, Pol II and Pol III transcripts, that contain 2 or more non-templated As.
(E) Transcriptome-wide binding profiles. Bar diagrams illustrate the percentage of all sequences mapped to the functional RNA classes indicated on the right of the figure.