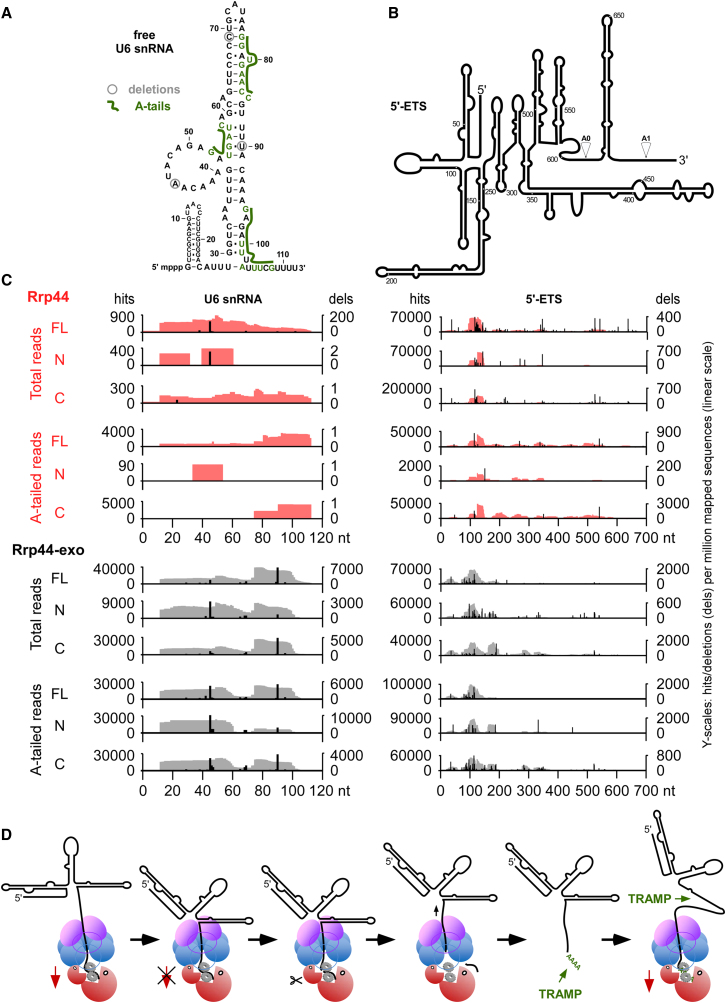
Figure 4.
Comparison of Rrp44 Domains in Split-CRAC
(A) Secondary structure of U6 snRNA (112 nt) from S. cerevisiae. Major sites of microdeletions are circled. These are due to reverse transcriptase stops at the crosslinked nucleotide. Prominent sites of oligoadenylation are indicated in green and are located 3′ to the major crosslinking sites. The positions of the first non-templated adenosines in A-tailed reads are indicated in green.
(B) Predicted secondary structure for the pre-rRNA 5′-ETS region (699 nt) from S. cerevisiae. Processing sites A0 and A1 are indicated by arrowheads.
(C) Read coverage for full-length and cleaved Rrp44 (red) and Rrp44-exo (black) in the U6 snRNA (left) and the pre-rRNA 5′-ETS region (right). Mapped reads (hits) are depicted in red (Rrp44) or gray (Rrp44-exo); positions of microdeletions (dels) are indicated in black. Data sets used for analysis were either unfiltered (Total) or filtered for reads containing 2 or more non-templated As (A-tailed). FL – full-length protein; N – NTD; C – CTD.
(D) Proposed model for the cooperative action of the endonuclease and exonuclease activities of Rrp44 and the TRAMP complex on structured RNA substrates. Many substrates are threaded through the exosome barrel to reach the active sites of Rrp44, which interacts with the exosome core via the NTD (Bonneau et al., 2009; Malet et al., 2010; Schaeffer et al., 2009; Schneider et al., 2009). Proteins of the RNase II family, which includes the exonuclease domain of Rrp44, are strongly processive and bind substrates tightly in the active site cleft (Zuo et al., 2006). This presumably allows Rrp44 to actively pull substrate RNAs in through the complex (1). However, only single stranded RNAs can enter the lumen of the exosome, so stable RNA-RNA or RNA-protein structures in the substrate potentially lead to stalled complexes, in which the 3′ end is tightly but non-productively bound by Rrp44 (2). We postulate that under these circumstances, the PIN domain cleaves the RNA (3), allowing substrate release (4). The substrate could then be re-adenylated by the TRAMP complex (5) and reloaded into the exosome (6), probably with the assistance of TRAMP.