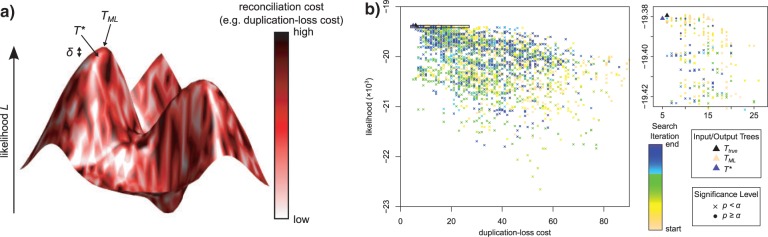
Figure 1.
Gene tree landscape. a) Each point within the landscape corresponds to a gene tree Tx, whose optimality can be measured through its likelihood Lx (height) and its reconciliation cost cx (color). The ML tree TML is located at the peak of this landscape but may have a high cost. Rearranging TML to a nearby tree Tx can result in a negligible decrease in likelihood (δx = LML − Lx < δthr) while simultaneously reducing the tree cost (cx < cML), thus producing a more congruent gene tree that is statistically equivalent to the ML tree. TreeFix utilizes this basic idea by balancing the 2 optimality criteria to return an optimal tree T* for which δ* is negligible and c* is minimal. b) The landscape for a simulated gene family shows a wide range of likelihood and cost values. In this instance, TreeFix searched over 3560 gene trees of the 8.2 × 1021 possible unrooted topologies (number of genes = 21). Although most trees have statistically worse likelihoods compared with the ML tree (×), a subset of high likelihood trees are statistically equivalent (circles). As the search progresses, the search space generally moves toward the top-left, corresponding to topologies with high likelihood and low duplication–loss cost (enlarged at right, accepted trees per iteration shown as squares). In this case, TreeFix has rearranged TML (beige triangle) to produce a new optimal tree T* (purple triangle) with equivalent likelihood and lower cost. Note that T* is incorrect because the true tree Ttrue (black triangle) has a slightly higher duplication–loss cost. (Likelihoods were computed with ϵ = 2.)