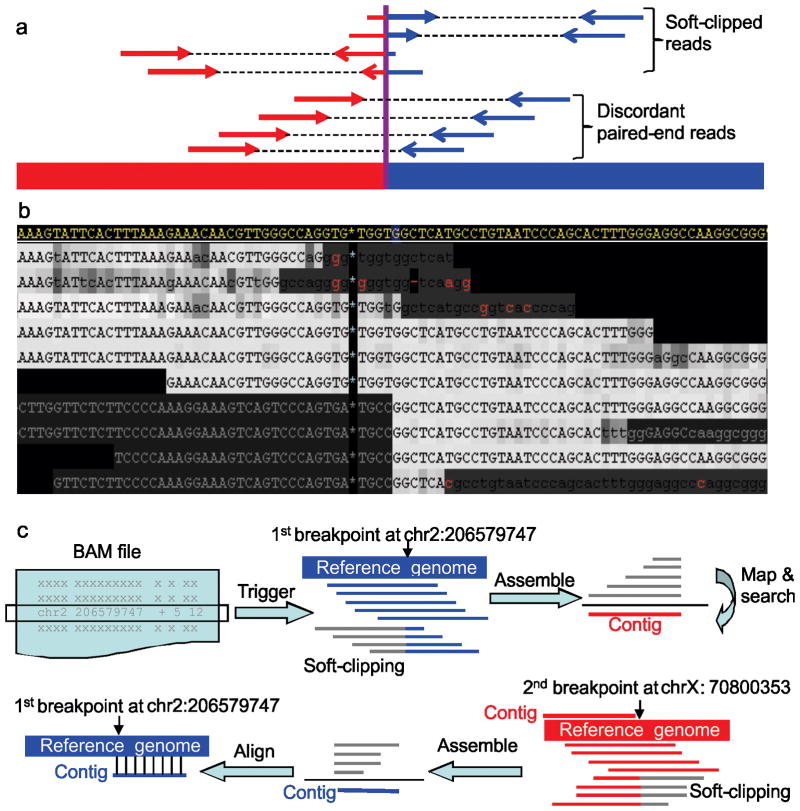
Figure 1.
Mapping SV breakpoints using soft-clipped reads. (a) Illustration of SV analysis using discordantly mapped paired-end reads versus mapping using soft-clipping reads. Red and blue segments represent two discontinuous genomic regions. (b) An example of using soft-clipping signature to identify an interchromosomal translocation. The region in display is 206579705-206579789bp on chromosome 2. The reference genome is shown at the top in yellow while the NGS reads are displayed below. Mismatches to the reference are shown in red letters while the gray letters at lower-left are soft-clipping subsequences not aligned to the reference. Upper-case characters represent high-quality (phred score ≥20) bases and the darkness of shading correlates to lower quality score. In this example, the soft-clipping subsequences map to chromosome X, revealing a chromosome 2 to chromosome X translocation. (c) The five-step CREST algorithm: extraction of soft-clipped reads (gray) in the BAM file; assembly of soft-clipped reads at a putative breakpoint into a contig (red); mapping of the contig against the reference genome to identify candidate partner breakpoints; identification of all possible soft-clipped reads and assembly into a contig (blue); and alignment of the contig derived from the partner (blue) back to the reference genome. A match to the initial breakpoint is considered a SV.