Figure 5.
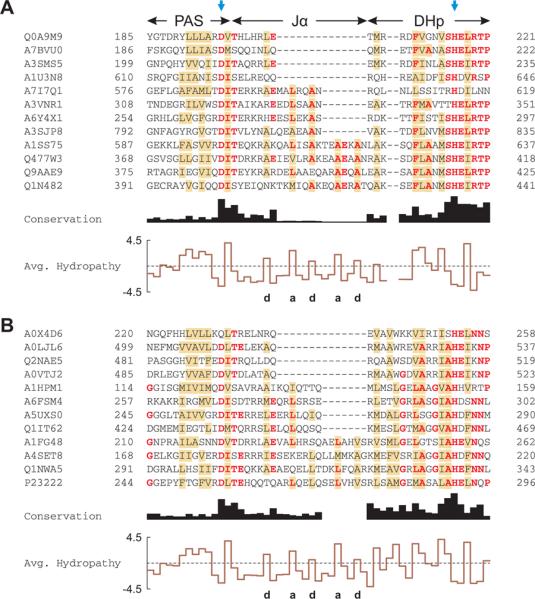
Multiple sequence alignment of PAS histidine kinases.
A. Alignment of PAS histidine kinases that have 7n residues between the C-terminus of the PAS domain and the active-site histidine, indicated by the blue arrows. Out of 1811 sequences analyzed, 12 are shown and labeled with their UniProt identifiers. Residues conserved in more than half of all 1811 sequences are shown in bold red; brown shading denotes columns with more than 50% hydrophobic residues. The C-terminus of the PAS domains displays a highly conserved DIT consensus motif. The linker between the PAS and DHp domains shows the pattern of hydrophobic residues (labeled a and d) characteristic of α-helical coiled coils. Two gap positions have been arbitrarily inserted into the alignment to facilitate comparison with panel B.
B. Alignment of PAS histidine kinases that have 7n+2 residues between the PAS domain and the active-site histidine. Out of 960 sequences analyzed, 12 are shown; B. japonicum FixL is in the bottom row (P23222). In comparison to panel A, the DHp domain differs in sequence and length around the phosphoacceptor histidine.
