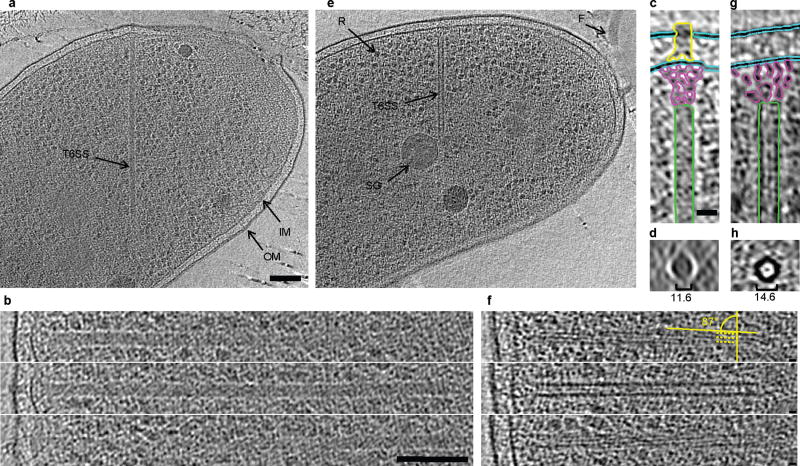
Figure 2. Electron cryotomographic imaging of T6SS structures inside intact cells.
Shown are different tomographic slices (19 nm in a, e, c, g; 9.5 nm in b,f; 190 nm in d, h) of an extended (a–d) and a contracted (e–h) structure imaged in two different wild type cells (contracted/extended structures, T6SS; IM, inner membrane; OM, outer membrane; F, flagellum; R, putative ribosome; SG, polyphosphate storage granule). (b) and (f), each show three slices at the same orientation but at different Z-heights. Compared to extended structures, contracted structures are shorter (b, f), have a helical surface pattern (pitch angle of 87°) and a smaller diameter (indicated in the perpendicular views in d, h). (c) and (g) are segmentations of densities observed in the extended (c) and contracted (g) structures. Densities shown in (h) originate from a contracted structure from a different tomogram. Segmented are putative densities corresponding to sheath (green), baseplate (pink and yellow) and membranes (blue). Bar in (a) 100 nm (applies to a, e), bar in (b) 100 nm (applies to b, f), bar in (c) 20 nm (applies to c, d, g, h).