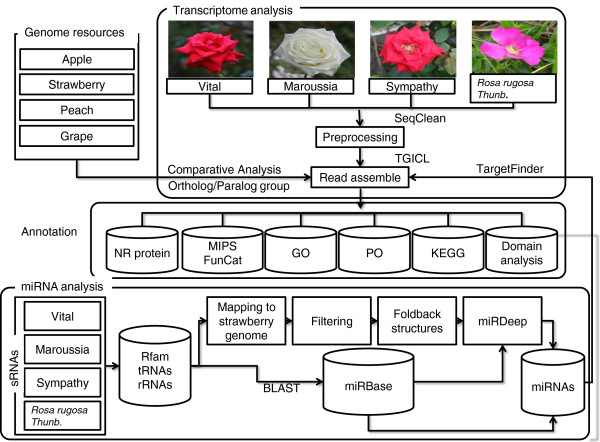
Figure 1.
Scheme for construction of the Rose database. The schematic presentation shows the strategy of transcriptome and miRNA analysis. Transcript reads were assembled by TGICL [32] after pre-processing with SeqClean. Assembled sequences were annotated by searching homologs against non-redundant (NR) database and Arabidopsis (ver. 10). Domains and Gene Ontology (GO) were analyzed by Pfam. KEGG metabolic pathways and plant ontology (PO) were assigned by referring the description of top matched Arabidopsis genes. miRNAs were identified by applying modified miRDeep [24,29]. To search for known miRNAs, sRNA sequences were BLAST searched against currently known miRNAs in miRBase [86] databases.