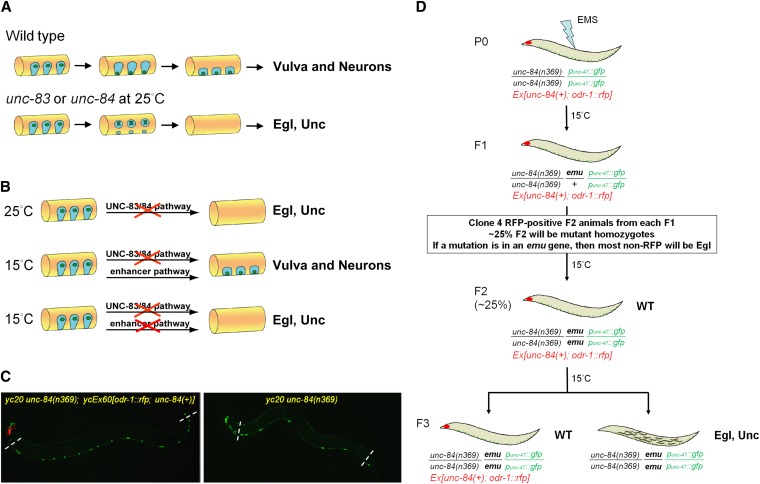
Figure 1 .
P-cell nuclear migration is temperature sensitive in an unc-83 or unc-84 background. (A) A conceptual diagram of the migration of nuclei (green) in P cells (blue) during the mid-L1 larval stage. A lateral view of the mid-body of an L1 animal (orange) is shown; ventral is down. In wild type, P-cell nuclei migrate from a lateral position to the ventral cord; the cytoplasm retracts following the nuclear migration. After migration, P-cell lineages normally develop into ventral neurons and vulva cells. In unc-83 or unc-84 null, mutant animals, failure of P-cell nuclear migration at 25° causes the death of many P cells and leads to egg-laying–deficient (Egl) and uncoordinated (Unc) phenotypes. (B) Our central hypothesis for an enhancer pathway mediating nuclear migration is shown. (Top line) At 25°, P-cell nuclear migration fails in unc-83 or unc-84 null animals. (Middle line) However, at 15°, unc-83 or unc-84 null animals behave normally, suggesting that a second, uncharacterized pathway (the enhancer or emu pathway) is sufficient for nuclear migration at 15°. (Bottom line) When both the unc-83/84 and emu pathways are disrupted, we hypothesized that the resulting animals would be Egl and Unc. (C) punc-47::GFP-labeled GABA neurons were used as a marker of P-cell–derived lineage. On the left, in yc20 unc-84(n369) animals with the unc-84–rescuing extrachromosomal array ycEx60[odr-1::rfp; unc-84(+)], 19 neurons were counted in the ventral cord between the pharynx and the anus (marked by dashed lines). However, failure of P-cell nuclear migration caused loss of GABA neurons as shown on the right where only 8 neurons were seen in a yc20 unc-84(n369) animal. Anterior is left. (D) An F2 clonal screen for emu in unc-84(n369) animals was performed using an extrachromosomal array ycEx60[odr-1::rfp; unc-84(+)] containing an unc-84(+) rescuing construct (see Materials and Methods for details).