Abstract
Background
The mammary gland is key to all mammal species; in particular in multiparous species like pigs the number and the shape of functional mammary gland complexes are major determinants of fitness. Accordingly, we aimed to catalog the genes relevant to mammogenesis in pigs. Moreover, we aimed to address the hypothesis that the extent and timing of proliferation, differentiation, and maturation proccesses during prenatal development contribute to postnatal numerical, morphological and functional properties of the mammary gland. Thus we focused on differentially expressed genes and networks relevant to mammary complex development in two breeds that are subject to different selection pressure on number, shape and function of teats and show largely different prevalence of non-functional inverted teats. The expression patterns of fetal mammary complexes obtained at 63 and 91 days post conception (dpc) from German Landrace (GL) and Pietrain (PI) were analyzed by Affymetrix GeneChip Porcine Genome Arrays.
Results
The expression of 11,731 probe sets was analysed between the two stages within and among breeds. The analysis showed the largest distinction of samples of the breed GL at 63 dpc from all other samples. According to Ingenuity Pathways Analysis transcripts with abundance at the four comparisons made (GL63-GL91, PI63-PI93, GL63-PI63 and GL91-PI91) were predominantly assigned to biofunctions relevant to `cell maintenance, proliferation, differentiation and replacement´, `organismal, organ and tissue development´ and `genetic information and nucleic acid processing´. Moreover, these transcripts almost exclusively belong to canonical pathways related to signaling rather than metabolic pathways. The accumulation of transcripts that are up-regulated in GL compared to PI indicate a higher proliferating activity in GL, whereas processes related to differentiation, maturation and maintenance of cells are more prominent in PI. Differential expression was validated by quantitative RT-PCR of five genes (GAB1, MAPK9, PIK3C2B, PIK3C3 and PRKCH) that are involved in several relevant signaling pathways.
Conclusions
The results indicate that mammary complex development in PI precedes GL. The differential expression between the two breeds at fetal stages likely reflects the prenatal initiation of postnatal phenotypes concerning the number and shape as well as functionality of teats.
Background
The development of the mammary gland is initiated during fetal stage. In the pig, the first visible structure at embryonic day 23 to 28 (E23 to E28) are elevated epidermal ridges or milk lines which are extending between forelimb to hindlimb on each side of trunk. The milk lines are a thickening of the ectoderm or the epidermis which are then fragment into individual buds. The formation of mammary placodes appears along each side of the body. In between E28 and E40, the placodes develop into bulb-shaped buds of epithelial cells by invagination into the underlying mesenchyme. Subsequently the size of the buds is slowly increasing and at E80 the mesenchymal cells surrounding the epithelial buds start to condense to become the mammary mesenchyme. Only late in prenatal development the epithelial buds elongate to the mammary mesenchyme to form a sprout, which creates a small duct. The sprout penetrates through fat pads. It starts to the ductal elongation and side branching about 10–15 times to form a rudimentary ductal tree. The mammary glands remain at this rudimentary stage, while the epithelial duct slowly grows until it reaches puberty [1-5].
Essentially, the development of mammary gland depends on growth hormones and growth factors. Moreover, the mammary gland development at fetal stages is apparently autonomous. The initiation of the mammary gland development and the early stage of morphogenesis are controlled by reciprocal interaction between epithelial and surrounding mesenchymal cells. The differentiation of mammary epithelia is also induced by the mammary mesenchyme [6-8]. During the differentiation at fetal stages the fate of cells towards their specialization as member of a population of cells typical for a tissue or organ is programmed. Accordingly, fetal development has implications on postnatal phenotypes. The mammary gland is key to all mammal species; in multiparous pigs the number and the shape of functional mammary gland complexes are major determinants of the mothering ability of sows.
In order to catalog genes relevant to mammogenesis in pigs, we analyzed the transcriptome of the mammary buds at the phase of formation and growth, i.e. at 63 day post conception (dpc) and 91 dpc, when epithelial and mesenchymal cell undergo proliferation and differentiation processes.
In order to address the hypothesis that balancing of proliferation and differentiation of epithelial and mesenchymal cells during prenatal development contributes to the postnatal shape and functionality of the mammary gland we compared fetal specimens obtained from two divergent breeds, German Landrace and Pietrain. Whereas in dam lines like German Landrace young sows are strongly selected for numerical, morphological and functional properties of the mammary gland, in sire lines like Pietrain these are not obligatory selection criterion. Accordingly, mammary complexes of both breeds differ in terms of number of teats and their distribution along the body and their symmetry at bold sides as well as the occurrence of additional non-functional teats including inverted teats. In fact, the examination of teat complexes of more than 2000 carcasses at the abattoir revealed mean teat numbers of around 15 and 13 in German Landrace and Pietrain, respectively [9], own observations]. The breeds also differ in the number of non-functional teats; interestingly, we observed different incidences of inverted teats of 12% in dam lines and 48% in Pietrain [10,11]. Thus differences of expression profiles of prenatal teat tissues between the two breeds could be indicative for molecular routes impertinent for the postnatal morphology and functionality of the mammary complexes.
The inverted teat defect is a polygenetic inherited liability trait marked by a decrease the number of functional teats thus causing animal welfare concerns due to increased piglet mortality and incidence of mastitis. It has been suggested that impaired prenatal development contributes to the emerging of inverted teats [10,12]. Moreover, we have previously demonstrated that genes of growth factor signaling pathways show differential expression depending on the teat phenotype and the affection status of the individual [11,13]. We aimed to elucidate whether molecular routes found to be affected due to the development of inverted teats at postnatal stages may already play a role during prenatal development. However, at prenatal stages the phenotype of inverted teat is not yet visible and the development of the teats cannot be predicted. In general, beside clones of animals that express a certain trait that is only visible during postnatal life but that is initiated by prenatal events, fetal samples of breeds that largely differ in that trait are the best available model.
The study was conducted with commercial genome-wide microarray (Affymetrix GeneChip Porcine Genome Array) to identify the differentially expressed genes and networks relevant to the development of mammary complexes in the two breeds and contributes to the understanding of molecular routes relevant to postnatal emergence of differences in morphological and functional properties.
Results
Gene expression
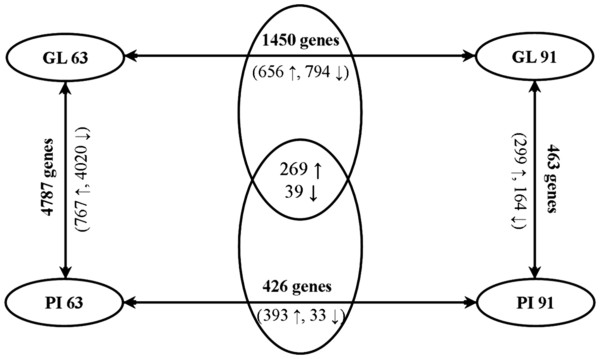
In order to identify differential gene expression in mammary gland development depending on stage (63 dpc and 91 dpc) and breed (GL and PI) probe sets (subsequently also referred to as genes or transcripts) with present calls in at least 50% of the samples were selected for statistical analysis summing up to 11,731 probe sets. Differential expression was evaluated between breeds within stages. Further we listed differentially expressed genes (DE-genes) between stages within breed. These two lists of DE-genes were compared. The commonly temporally regulated genes were considered separate from those genes that were specific to either of the two breeds because only the later reflect breed differences, whereas commonly regulated genes between stages in both breeds are not likely to contribute to the initiation of divergence of the postnatal phenotype among the breeds. The numbers of significant differentially expressed genes are summarized in Figure 1. Fold changes (FC) varied between 1.2 and 50.2. Most pronounced differences were found between breeds at stage 63 dpc: at p < 0.05 corresponding to q = 0.005-0.03 there were 4787 DE-genes with median FC of 2.1 and a maximum FC of 50.2. Also between stages within the breed GL considerable transcriptomic differences were obvious (number of DE-genes: 1758 at p < 0.05 corresponding to q = 0.001-0.14; FCmedian = 1.7; FCmax = 19.2; number of DE-genes temporally regulated but specific to GL: 1450 at p < 0.05 corresponding to q = 0.001-0.14; FCmedian = 1.8; FCmax = 11.4), whereas between stages 63 dpc and 91 dpc in PI (number of DE-genes: 734 at p < 0.05 corresponding to q = 0.04-0.8; FCmedian = 1.6; FCmax = 25.4; number of DE-genes temporally regulated but specific to PI: 426 genes at p < 0.05 corresponding to q = 0.04-0.8; FCmedian = 1.7; FCmax = 19.7) and among GL and PI at 91 dpc (number of DE-genes: 463 at p < 0.05 corresponding to q = 0.5; FCmedian = 1.7; FCmax = 11.9) the distinctness of the expression profiles was moderate.
Figure 1.
The number of significant differentially expressed genes in same breeds or stages and different breeds or stages. in bold: numbers of transcripts with difference abundance at the respective comparisons (GL63-GL91, PI63-PI93, GL63-PI63 and GL91-PI91) at p < 0.05. in parenthesis: number of transcripts with higher or lower abundance relative to the breed PI (↑ = PI higher or ↓ = PI lower)during development from 63 dpc to 91 dpc (↑ = increase, ↓ = decrease).
Ingenuity pathway analysis
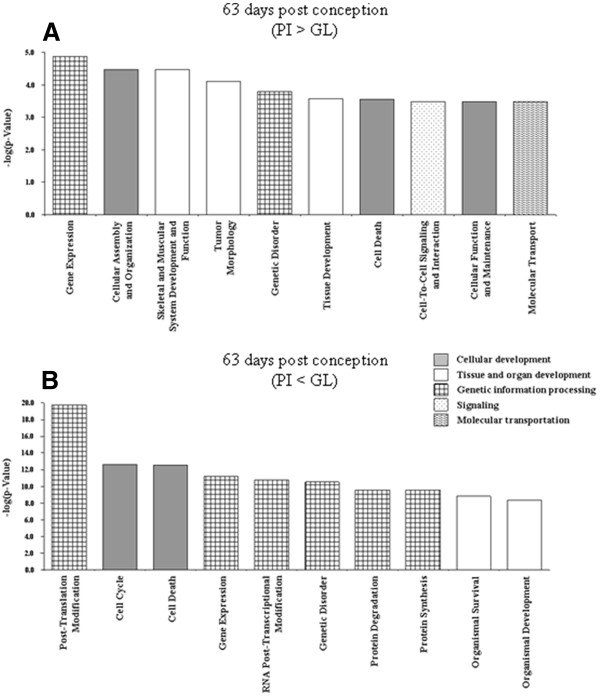
Four lists of DE-genes were imported to the Ingenuity Pathways Analysis (IPA) in order to assign them to groups or categories of biofunctions and to canonical pathways as defined in the Ingenuity Knowledge Base (Genes) and to test for significant enrichment of DE-genes within these groups. The DE-genes were predominantly assigned to biofunctions relevant to `cell maintenance, proliferation, differentiation and replacement´, `organismal, organ and tissue development´ and `genetic information and nucleic acid processing´. Referring to the most significant functions named by IPA within the IPA-categories listed in Figures 2, 3, 4, 5 elucidates the developmental status of the mammary gland tissues within the four groups compared here. Those genes that are higher expressed in PI than in GL at 63 dpc were assigned to genetic information processing, in particular transcription and transactivation (IPA-category: `gene expression´), cellular processes including formation of filaments and intercellular junction (IPA-category: `cellular assembly and organization´), survival of cells (IPA-category: `cell death), and cytostasis (IPA-category: `cellular function and maintenance´)(Figure 2A). At 63 dpc in DL many genes annotated to biofunctions related to genetic information and nucleic acid processing are higher expressed than in PI; this covers functions of transcription (IPA-category: `gene expression´), RNA processing and splicing (IPA-category: `RNA post-transcriptional modification´), apoptosis (IPA-category: `cell death´) but also cell division (IPA-category: `cell cycle´) and also includes genes relevant to early onset of breast cancer (IPA-category: `genetic disorder´) (Figure 2B). The accumulation of DE-genes that are up-regulated in DL compared to PI indicate a higher proliferating activity in DL, whereas processes related to maintenance and differentiation and maturation of cells are more prominent in PI at 63 dpc.
Figure 2.
Significant biofunctions (top ten according to p-value) representing genes differentially expressed between samples of mammary complexes of Pietrain and German Landrace at 63 dpc.(A) PI > GL (B) PI < GL. All assignments significant after Benjamini–Hochberg correction, except `tissue development´, `cell death´, cell-to-cell signaling´, `cellular function and maintenance´, and `molecular transport´ in (A).
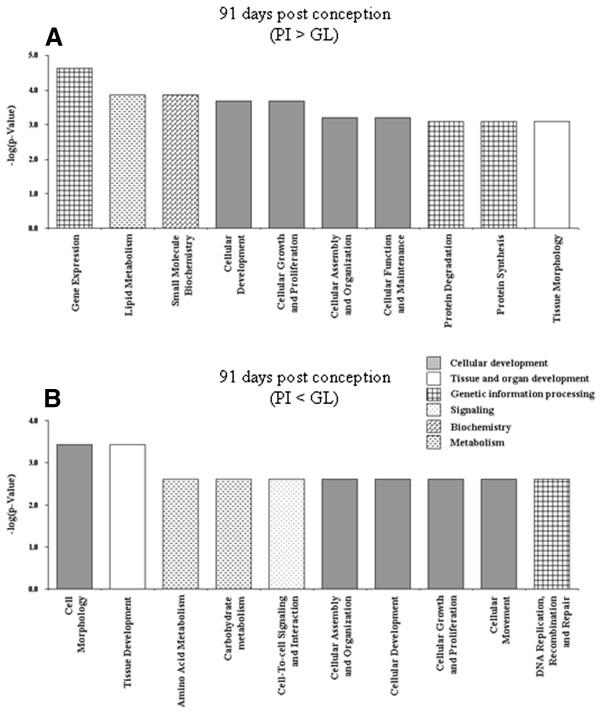
Figure 3.
Significant biofunctions (top ten according to p-value) representing genes differentially expressed between samples of mammary complexes of Pietrain and German Landrace at 91 dpc.(A) PI > GL (B) PI < GL. All assignments significant after Benjamini–Hochberg correction.
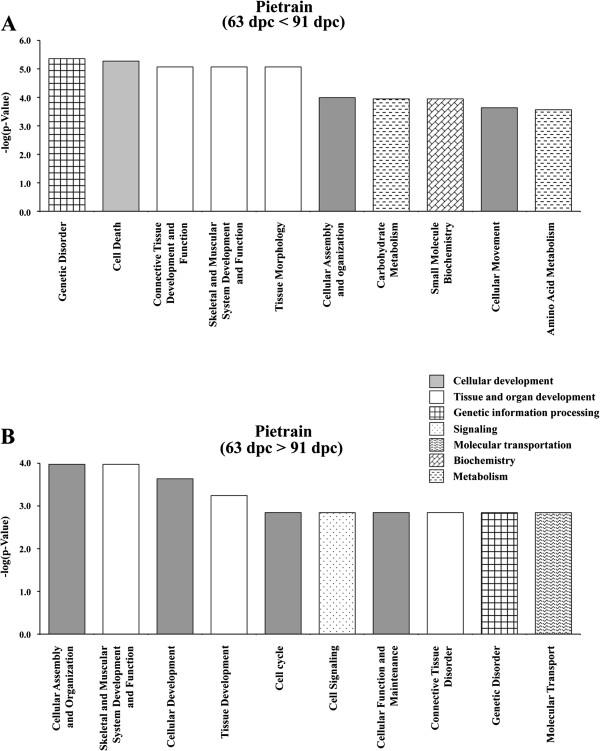
Figure 4.
Significant biofunctions (top ten according to p-value) representing genes differentially expressed in Pietrain at 63 dpc compared to 91 dpc.(A) 63 dpc < 91 dpc (B) 63 dpc > 91 dpc. All assignments significant after Benjamini–Hochberg correction.
Figure 5.
Significant biofunctions (top ten according to p-value) representing genes differentially expressed in German Landrace at 63 dpc compared to 91 dpc.(A) 63 dpc < 91 dpc (B) 63 dpc > 91 dpc. All assignments significant after Benjamini–Hochberg correction.
At 91 dpc biofunctions related to cellular development are predominantly regulated in both breeds (Figure 3A, B). In GL genes with function in cell differentiation including the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state (IPA-category: `cellular development´) as well as stem cell proliferation (IPA-category: `cellular growth and proliferation´) are up-regulated compared to PI (Figure 3B). Cell maturation, i.e. developmental processes, independent of morphogenetic (shape) change, that are required for a cell to attain its fully functional state (IPA-category: `cellular development´) and growth (IPA-category: `cellular growth and proliferation´) are biofunctions covering a significant number of genes higher expressed in PI than in GL at 91 dpc (Figure 3A). Thus also at 91 dpc the results suggest that the tissue of the mammary complex has already reached a higher degree of maturity in PI than GL. However, none of the transcripts considered here in this comparison among breeds at 91 dpc reached a false discovery rate below 0.5.
Corresponding to the differential regulation between breeds within stages the temporal regulation between 63 dpc and 91 dpc that is limited to PI indicates a high expression of genes related to differentiation and maintenance of cell, i.e. survival of cells (IPA-category: `cell death´), organization of organelles and cytoskeleton (IPA-category: `cellular assembly and organisation´) and remodelling of tissue and quantity of connective tissue cells (IPA-categories: `connective tissue development and function´ as well as `tissue morphology´) (Figure 4). When only taking into account transcripts with different abundance at p < 0.001 (corresponding to q < 0.27) 44 remain that do not provide a meaningful IPA. The temporally regulated genes that are restricted to the breed GL were assigned to processes impertinent to dynamic changes of the cell population within the tissue at 63 dpc with functions like proliferation of cells (IPA-category: `cellular growth and proliferation´), transcription (IPA-category: `gene expression´), apoptosis (IPA-category: `cell death´), and cell migration (IPA-category: `cellular movement´) (Figure 5). Assignment to biofunctions of commonly regulated genes between stages in both breeds is shown in the supplementary material (Additional file 1: Figure S1).
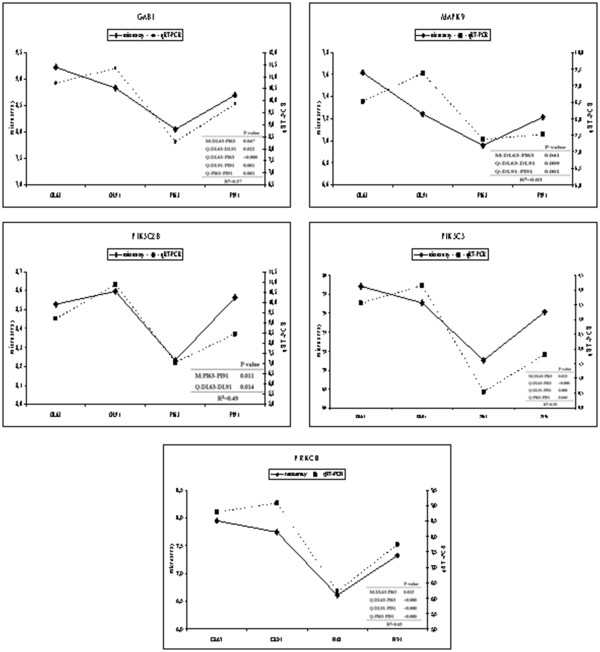
IPA towards assignment of DE-genes to canonical pathways pointed to signaling pathways important for cellular proliferation, differentiation, development and growth as detailed in Tables 1 and 2. We selected five genes (GAB1, MAPK9, PIK3C2B, PIK3C3 and PRKCH) that are involved in several signaling pathways and therefore were redundantly listed (in bold in Tables 1 and 2) for examination of their expression by qRT-PCR (Figure 6). In general, the breed and stage dependent differences of relative transcript abundance as found by microarray analysis and qRT-PCR were in good agreement (Figure 6). The relative transcript abundance according to microarrays and qRT-PCR were significantly correlated at R² = 0.49 - 0.65, except for MAPK9. For MAPK9 microarrays indicated highest expression GL at 63 dpc, whereas qRT-PCR indicated an even higher expression in GL at 91 dpc; however in both analyses expression in PI was lower than in GL at any of the two stages.
Table 1.
Assignment of temporally regulated DE-genes to canonical pathways in German Landrace and Pietrain, respectively
| Ingenuity Canonical Pathways | 63 dpc vs. 91 dpc in GL or PI | p-value1 | Ratio2 | Molecules |
|---|---|---|---|---|
| Angiopoietin Signaling |
GL |
0.008 |
0.143 |
STAT5A, PAK4, AKT2, IKBKG, TNIP1, GRB14, ANGPT1, PAK6, PTPN11, MRAS, IKBKAP |
| PI |
0.065 |
0.065 |
PIK3C2B, PAK4, IKBKG, TNIP1, PIK3R4 |
|
| Chemokine Signaling |
GL |
0.028 |
0.133 |
GNAI3, PLCB4, JUN, MYL2, CXCL12, MRAS, GNAQ, PPP1R12A, MAPK12, CAMK2G |
| PI |
0.393 |
0.040 |
CAMK2A, MYL2, CAMK2G |
|
| ILK Signaling |
GL |
0.005 |
0.120 |
MYH10, AKT2, VEGFB, MYL2, TNFRSF1A, CREB3, MYH11, MAPK12, MYL9, NCK2, RHOQ, JUN, RHOG, IRS1, RHOU, CHD1, MYH9, LEF1, ACTG2, PTGS2, PPP2R5E, ACTC1, ACTN1 |
| PI |
0.002 |
0.073 |
PIK3C2B, MYL2, TNFRSF1A, VEGFC, MYH11, PIK3R4, MYL6B, PPP2R5A, ITGB3, PPP2R4, PPP2R2B, MYH3, PTGS2, ACTC1 |
|
| Integrin Signaling |
GL |
0.011 |
0.112 |
CAPN5, FYN, AKT2, PAK4, ARHGAP26, TSPAN7, MYL2, PAK6, ARHGEF7, TNK2, MYLK, NCK2, RHOG, RHOQ, MRAS, RHOU, PPP1R12A, ITGA1, ACTG2, ACTC1, ACTN1, RAP2A, RAPGEF1 |
| PI |
0.275 |
0.039 |
PIK3C2B, PAK4, MYL2, TSPAN7, PIK3R4, ACTC1, TTN, ITGB3 |
|
| TGF-β Signaling |
GL |
0.008 |
0.145 |
SMAD2, BMPR1B, JUN, SMAD9, TGFB1, MRAS, TGFB3, TGFB2, SMAD6, PITX2, SMAD1, INHBB |
| PI |
0.228 |
0.048 |
SMAD2, TGFB1, TGFB3, INHBB |
|
| Wnt/β-catenin Signaling |
GL |
0.019 |
0.114 |
AKT2, SFRP2, APPL2, PPARD, WNT2B, MARK2, GNAQ, APPL1, CDH2, JUN, TGFB1, CSNK2A1, TGFB2, TGFB3, NR5A2, FZD5, LEF1, SFRP1, PPP2R5E, TCF7L2 |
| PI |
0.046 |
0.057 |
CDH2, CDH1, PPP2R4, TGFB1, PPP2R2B, CD44, TGFB3, FZD6, TLE1, PPP2R5A |
|
| Cyclins and Cell Cycle Regulation |
GL |
0.023 |
0.124 |
CCND3, PA2G4, HDAC8, TGFB1, E2F1, HDAC7, E2F5, TGFB3, TGFB2, PPP2R5E, SKP2 |
| PI |
0.003 |
0.090 |
CCNE2, PPP2R4, TGFB1, PPP2R2B, TGFB3, CCNB2, ATR, PPP2R5A |
|
| Aryl Hydrocarbon Receptor Signaling |
GL |
0.007 |
0.116 |
ALDH4A1, NFIC, MED1, HSPB2, CYP1B1, CTSD, ALDH1A1, JUN, CCND3, NCOA2, TGFB1, E2F1, TGFB3, TGFB2, IL1B, NFIB, NCOR2, ESR1 |
| PI |
0.004 |
0.071 |
ALDH4A1, CCNE2, ALDH1A1, TGFB1, TGFB3, IL1B, ALDH18A1, NCOR2, ATR, ESR1, NCOA3 |
|
| Gα12/13 Signaling |
GL |
0.043 |
0.109 |
F2RL2, AKT2, F2R, MYL2, MAPK12, LPAR3, CDH11, MYL9, CDH2, IKBKG, JUN, LPAR1, MRAS, MAPK7 |
| PI |
0.005 |
0.078 |
PIK3C2B, CDH2, CDH1, IKBKG, MYL2, MEF2C, MAP3K5, MAPK7, PIK3R4, MYL6B |
|
| Glucocorticoid Receptor Signaling |
GL |
0.025 |
0.095 |
ICAM1, GTF2A2, TSC22D3, IKBKG, JUN, NCOA2, TGFB1, MRAS, TGFB2, GTF2H5, POLR2H, NCOR1, FKBP5, TAF12, SMAD2, STAT5A, AKT2, MED1, TAF15, CEBPB, MAPK12, NCOA1, TGFB3, IL1B, PTGS2, NCOR2,ESR1 |
| PI |
0.033 |
0.049 |
SMAD2, PIK3C2B, SMARCD2, CEBPB, PIK3R4, NCOA3, TSC22D3, IKBKG, TGFB1, TGFB3, IL1B, NCOR2, PTGS2, ESR1 |
|
| Ceramide Signaling |
GL |
0.352 |
0.078 |
CTSD, AKT2, JUN, TNFRSF1A, MRAS, PPP2R5E, TNFRSF1B |
| PI |
0.044 |
0.067 |
PIK3C2B, PPP2R4, TNFRSF1A, PPP2R2B, PIK3R4, PPP2R5A |
|
| PPARα/RXRα Activation |
GL |
0.002 |
0.123 |
PPARA, SMAD2, MED1, ADCY3, GNAQ, ADCY6, MAP4K4, CAND1, PLCD1, PLCD3, IKBKG, PLCB4, JUN, TGFB1, FASN, IRS1, MRAS, TGFB3, TGFB2, IL1B, NCOR1, NCOR2, INSR |
| PI |
0.020 |
0.059 |
PLCD1, SMAD2, IKBKG, TGFB1, GNA11, ADCY6, TGFB3, IL1B, NCOR2, NCOA3, ABCA1 |
|
| RhoA Signaling |
GL |
0.019 |
0.127 |
MYL2, RDX, WASF1, DLC1, LPAR3, MYLK, MYL9, LPAR1, IGF1R, PPP1R12A, CDC42EP1, ACTG2, ACTC1, PI4KA |
| PI |
0.007 |
0.082 |
RHPN2, IGF1, MYL2, EPHA1, IGF1R, ARHGAP12, MYL6B, ACTC1, TTN |
|
| Tight Junction Signaling |
GL |
0.004 |
0.127 |
MYH10, F2RL2, TIAM1, AKT2, MYL2, TNFRSF1A, PVRL3, MARK2, MYH11, CASK, MYL9, MYLK, JUN, TGFB1, TGFB3, TGFB2, MYH9, ACTG2, PPP2R5E, TNFRSF1B, ACTC1 |
| PI |
0.005 |
0.072 |
MYL2, CLDN8, PPP2R4, TGFB1, TNFRSF1A, PPP2R2B, MYH3, TGFB3, MYH11, MYL6B, ACTC1, PPP2R5A |
|
| Estrogen-Dependent Breast Cancer Signaling |
GL |
0.076 |
0.110 |
STAT5A, AKT2, JUN, CREB3, IGF1R, MRAS, HSD17B7, ESR1 |
| PI |
0.014 |
0.082 |
PIK3C2B, IGF1, IGF1R, PIK3R4, ESR1, HSD17B8 |
|
| Mitotic Roles of Polo-Like Kinase |
GL |
0.424 |
0.079 |
CDC25B, TGFB1, CDC23, PPP2R5E, ANAPC13 |
| PI |
0.012 |
0.095 |
PPP2R4, TGFB1, PPP2R2B, CCNB2, CDC27, PPP2R5A |
|
| PPAR Signaling |
GL |
0.000 |
0.170 |
PPARA, STAT5A, TNFRSF1A, PPARD, MED1, MAP4K4, IKBKG, JUN, NCOA1, MRAS, IL1B, NCOR1, PTGS2, NCOR2, INSR, TNFRSF1B, CITED2, PDGFRB |
| PI |
0.081 |
0.057 |
IKBKG, TNFRSF1A, IL1B, NCOR2, PTGS2, PDGFRB |
|
| Role of Tissue Factor in Cancer |
GL |
0.164 |
0.095 |
FYN, STAT5A, YES1, AKT2, PTPN11, MRAS, GNAQ, IL1B, GNA14, RPS6KA1, MAPK12 |
| PI |
0.009 |
0.078 |
PIK3C2B, CTGF, CSF1, GNA11, VEGFC, IL1B, RPS6KA1, PIK3R4, ITGB3 |
|
| TR/RXR Activation |
GL |
0.010 |
0.130 |
AKT2, NXPH2, MED1, THRA, KLF9, SCARB1, COL6A3, NCOA2, FASN, NCOA1, STRBP, NCOR1, NCOR2 |
| PI | 0.063 | 0.060 | KLF9, PIK3C2B, COL6A3, NCOR2, PIK3R4, NCOA3 |
1pathways are shown that were significant at p < 0.05 according to Fishers exact test in at minimum one of the three types of comparisons.
2ratio of number of differentially expression genes assigned to the pathway and the total number of genes assigned to the pathway in the Ingenuity Knowledge Base.
Table 2.
Assignment of DE-genes to canonical pathways in the comparison between breeds at either 63 dpc or 91 dpc
| Ingenuity Canonical Pathways | PI vs. GL at 63 dpc or 91 dpc | p-value1 | Ratio2 | Molecules | |
|---|---|---|---|---|---|
| Clathrin-mediated Endocytosis Signaling |
63 |
0.011 |
0.222 |
EPS15, STON2, CDC42, FGF2, ARPC5, NUMB, SH3GL2, ITGB8, PIK3R4, CD2AP, ACTR3, WASL, SNX9, IGF1, PIK3C3, ARPC3, DAB2, STAM, AAK1, PPP3CA, ACTA1, ATM, ITGB1, MYO6, ACTR2, PIK3C2A, SH3GL3, ACTB, CLTC, RAB7A, MET, CBL, SYNJ1, ARPC2, RAB11A, TFRC, UBC, PDGFD |
|
| 91 |
0.106 |
0.029 |
EPS15, FGF2, TFRC, DAB2, AAK1 |
||
| Corticotropin Releasing Hormone Signaling |
63 |
0.033 |
0.204 |
RAP1B, PRKACB, RAF1, MAPK1, ARPC5, CREB5, PRKAG1, PRKD3, PRKCA, null, ITPR2, CNR1, PTCH1, GNAQ, GNAI1, ADCY6, MAPK12, RAP1A, ATF2, GNAS, GNAI3, PRKCI, MAPK14, PRKAR2B, PRKAG2, PRKCH, GLI1, PRKCB |
|
| 91 |
0.258 |
0.022 |
PRKAR2B, ADCY3, PTGS2 |
||
| Integrin Signaling |
63 |
0.000 |
0.259 |
MAP2K4, RAP2B, RAF1, MYL2, MAPK1, ARPC5, ITGA8, KRAS, PIK3R4, PTEN, TSPAN3, RHOG, ARF4, CAV1, ITGAV, GSK3B, ACTA1, ATM, CAPN5, ACTR2, BCAR3, RAP1A, TTN, RHOQ, RND3, ARPC2, PPP1R12A, CAPN7, TSPAN6, RAP1B, FYN, PPP1CC, RALA, CDC42, PPP1CB, ITGB8, SHC1, ACTR3, WASL, RHOT1, PIK3C3, SOS1, ARPC3, ITGB1, PAK2, PIK3C2A, ASAP1, ACTB, ITGA2, MAPK8, ROCK1, WIPF1, ITGAX |
|
| |
91 |
0.191 |
0.024 |
RALA, ASAP1, ARF4, ITGA8, TTN |
|
| PI3K/AKT Signaling |
63 |
0.048 |
0.197 |
RAF1, MAPK1, INPPL1, KRAS, JAK2, MAP3K5, EIF4E, PTEN, BCL2, SHC1, IKBKG, SOS1, TSC2, GSK3B, MCL1, ITGB1, RPS6KB1, YWHAG, PPP2R5C, ITGA2, TYK2, YWHAZ, PPP2R5A, PPP2CB, GAB1, CDKN1B, PPP2R5E, PPP2R1B |
|
| 91 |
0.296 |
0.021 |
PTGS2, PPP2R5A, BCL2 |
||
| α-Adrenergic Signaling |
63 |
0.012 |
0.236 |
PRKACB, RAF1, MAPK1, GNB5, KRAS, PRKAG1, PHKA2, GNB1, GNB4, PHKB, PRKD3, PRKCA, null, ITPR2, GNAI1, ADCY6, GNAQ, GNAS, GNAI3, PRKCI, PRKAR2B, PRKAG2, PRKCH, GNG2, PRKCB |
|
| 91 |
0.404 |
0.019 |
PRKAR2B, ADCY3 |
||
| IL-15 Signaling |
63 |
0.047 |
0.229 |
STAT5A, RAF1, PIK3C2A, MAPK1, TYK2, KRAS, JAK2, AXL, MAPK12, PIK3R4, BCL2, SHC1, MAPK14, PIK3C3, SYK, ATM |
|
| 91 |
0.267 |
0.029 |
STAT6, BCL2 |
||
| Myc Mediated Apoptosis Signaling |
63 |
0.018 |
0.266 |
MAP2K4, YWHAG, PIK3C2A, MAPK8, YWHAZ, MAPK9, KRAS, PIK3R4, MAPK12, BCL2, SHC1, IGF1, PIK3C3, SOS1, CYCS, BID, ATM |
|
| 91 |
0.255 |
0.031 |
APAF1, BCL2 |
||
| Protein Kinase A Signaling |
63 |
0.000 |
0.228 |
PRKACB, MYH10, RAF1, TGFBR1, MAPK1, MYL2, PDE12, GNB5, AKAP3, CREB5, PPP1R14B, TGFBR2, GNB1, GNB4, PHKB, CAMK2A, TDP2, GSK3B, PRKD3, null, YWHAG, ITPR2, PTCH1, CREBBP, YWHAZ, RAP1A, MYL6B, TTN, ATF2, MYL9, AKAP13, ANAPC4, ANAPC5, PPP1R12A, PRKCH, LEF1, GNG2, PDE6D, AKAP12, RAP1B, PPP1CC, FLNB, PDE7A, AKAP8, PDIA3, PPP1CB, H3F3A/H3F3B, CDC23, PRKAG1, PHKA2, NFAT5, TGFB2, SMAD4, PPP3CA, PRKCA, AKAP5, ATF1, MAP3K1, ADCY6, GNAI1, GNAQ, ANAPC13, ROCK1, GNAS, GNAI3, PPP1R3D, PRKAR2B, PRKCI, ADD3, KDELR2, PRKAG2, AKAP9, PRKCB, ANAPC1 |
|
| 91 |
0.348 |
0.019 |
ADD3, PRKAR2B, ADCY3, AKAP3, AKAP7, TTN |
||
| Molecular Mechanisms of Cancer |
63 |
0.000 |
0.275 |
RAP2B,RAF1,TGFBR1,APH1B,TAB2,ARHGEF1,KRAS,RBL1,RB1,CAMK2A,HIPK2,PRKD3,ATM,SMAD2,TFDP1,PTCH1,CREBBP,RAP1A,CDH1,GAB1,E2F1,CYCS,CFLAR,RAP1B,FYN,RALA,CDC42,LRP6,BMPR2,CRK,JAK2,MAP3K5,GNA14,CHEK1,CASP6,PIK3C3,SOS1,E2F5,BID,BMP1,PAK2,GNAQ,ADCY6,MAPK8,GNAI3,RBPJ,ATR,BIRC2,PSEN1,PRKCB,MAP2K4,PRKACB,MAPK1,PIK3R4,TGFBR2,RHOG,GSK3B,RASA1,BIRC3,TYK2,CDK6,MAPK12,RALBP1,APC,RHOQ,CBL,PTPN11,RND3,FZD6,PRKCH,LEF1,FZD5,CDK2,HIF1A,E2F3,PRKAG1,BCL2,CDC25B,SHC1,FANCD2,RHOT1,BMPR1A,MAP3K7,TGFB2,SMAD4,PRKCA,ARHGEF12,PIK3C2A,HAT1,GNAI1,MAPK9,XIAP,GNAS,PRKCI,MAPK14,PRKAR2B,FZD4,NF1,BMP8B,PRKAG2,CDKN1B,GLI1,BCL2L11 |
|
| 91 |
0.017 |
0.029 |
PRKAR2B,RALA,FZD4,ADCY3,APAF1,RAPGEF3,E2F3,CASP7,E2F2,WNT5A,BCL2 |
||
| p53 Signaling |
63 |
0.000 |
0.293 |
GADD45G, PIK3R4, PTEN, CHEK1, BCL2, RB1, CASP6, GADD45A, PIK3C3, GSK3B, HIPK2, ATM, TP53INP1, TP63, PIK3C2A, TOPBP1, MED1, THBS1, HDAC1, PERP, MAPK8, TP53BP2, KAT2B, PCNA, MAPK14, E2F1, ATR, CDK2, SIRT1 |
|
| 91 |
0.192 |
0.030 |
MED1, APAF1, BCL2 |
||
| VDR/RXR Activation |
63 |
0.003 |
0.284 |
CYP24A1, SPP1, CCNC, MED1, IGFBP5, CEBPB, THBD, KLF4, NCOA3, GTF2B, PRKCI, SP1, GADD45A, NCOA2, MXD1, NCOA1, IGFBP3, TGFB2, PRKCH, CDKN1B, PRKD3, PRKCA, PRKCB |
|
| 91 |
0.124 |
0.037 |
CYP24A1, MED1, MXD1 |
||
| Breast Cancer Regulation by Stathmin1 |
63 |
0.000 |
0.248 |
PRKACB, RAF1, CAMK1D, MAPK1, GNB5, KRAS, ARHGEF1, PIK3R4, PPP1R14B, GNB1, GNB4, CAMK2A, PRKD3, ATM, null, ITPR2, PPP2CB, E2F1, PPP1R12A, PRKCH, GNG2, CDK2, PPP1CC, CDC42, PPP1CB, E2F3, PRKAG1, SHC1, PIK3C3, SOS1, RB1CC1, E2F5, PRKCA, ARHGEF12, PIK3C2A, PPP2R5C, GNAI1, TUBA4A, ADCY6, GNAQ, PPP2R5A, ROCK1, GNAS, GNAI3, PPP1R3D, PRKCI, PRKAR2B, PRKAG2, CDKN1B, PPP2R5E, PPP2R1B, PRKCB |
|
| 91 |
0.191 |
0.024 |
PRKAR2B, ADCY3, E2F3, E2F2, PPP2R5A |
||
| ERK/MAPK Signaling |
63 |
0.001 |
0.230 |
RAP1B, PRKACB, FYN, PPP1CC, RAF1, MAPK1, HSPB2, H3F3A/H3F3B, ETS2, PPP1CB, KRAS, CRK, PIK3R4, CREB5, PPP1R14B, EIF4E, PRKAG1, SHC1, PIK3C3, SOS1, MKNK1, PRKCA, ATM, ITGB1, MYCN, PAK2, YWHAG, PPP2R5C, ATF1, PIK3C2A, ITGA2, YWHAZ, MAPKAPK5, RAP1A, PPP2R5A, ATF2, PLA2G4A, PPP2CB, PPP1R3D, PRKCI, PRKAR2B, PRKAG2, PPP1R12A, PPP2R5E, PPP2R1B, ELK3, PRKCB |
|
| 91 |
0.556 |
0.015 |
PRKAR2B, RAPGEF3, PPP2R5A |
||
| RAR Activation |
63 |
0.001 |
0.246 |
MAP2K4, PRKACB, NSD1, MAPK1, MAP3K5, JAK2,RBP1, PRKAG1, PTEN, PNRC1, TGFB2, SMAD4, GTF2H5, NR2F6, RDH13, PRKD3, CITED2, PRKCA, STAT5A, SMAD2, SRA1, PRMT2, IL3RA, RDH14, MED1, RDH11, MAP3K1, CREBBP, MAPK8, ADCY6, MAPK9, MAPK12, CRABP1, PARP1, KAT2B, PRKCI, MAPK14, PRKAR2B, TAF4, ERCC3, IGFBP3, NCOA1, PRKAG2, PRKCH, PRKCB |
|
| 91 | 0.269 | 0.022 | PRKAR2B, MED1, ADCY3, RDH13 |
1pathways are shown that were significant at p < 0.05 according to Fishers exact test in at minimum one of the three types of comparisons.
2ratio of number of differentially expression genes assigned to the pathway and the total number of genes assigned to the pathway in the Ingenuity Knowledge Base.
Figure 6.
Comparison of microarray (left y-axis) and quantitative real time PCR (right y-axis) data of five genes. Graphs show log(2) mean values of transcript abundance of the breed categories `German Landrace´ (GL), and `Pietrain´ (PI) at the prenatal stage of 63 and 91 day post conception `GL63´, `GL91´, `PI63´ and `PI91´.
Discussion
Differential expression at bud formation and growth among divergent breeds
The mammary gland development begins during fetal development and proceeds through the adult life. The fetal mammary gland development in pigs initiates at day 23 (E23) with the formation of the milk lines and continues with the invagination of epithelial cells to form buds that subsequently increase in size accompanied by the condensation of surrounding mesenchymal cells. The mammary gland development at fetal stages depends on the signal between epithelium and surrounded mesenchyme. Epithelial and mesenchyme are found in every organ. Either the transitions of epithelial into mesenchyme or the transitions of mesenchyme into epithelial is the cause of morphogenesis, differentiation and growth during the development [14]. A mesenchymal-epithelial interaction is a reciprocal process, where mesenchyme induces the formation of epithelial, while epithelium induces mesenchymal differentiation [15,16]. In fetal mammary gland development, mammary mesenchyme induces the formation of mammary epithelial buds when they were combined with the epidermis. The mammary buds induce the expression of androgen receptors (AR) and tenascin in mesenchymal cells and then the formation of the primary mammary mesenchyme occurs [15]. The insufficient proliferation of mesenchyme in embryonic mammary development may cause the inverted teat [13].We analysed samples 63 dpc and 91 dpc during growth of buds and their protrusion by mesenchymal cell development that is important for the postnatal shape and function of teats. Furthermore, hypothesising that the prenatal development of the buds affects the postnatal phenotype of teats we compared samples of two breeds that are subject to different selection pressure on number, shape and function of teats. The analysis of differential expression in the fetal tissues at the two time points between the two breeds showed the largest distinction of samples of the breed GL at 63 dpc from all other samples. Accordingly, the identification of transcripts with different abundance in specimens of GL at 63 dpc compared to those of PI at the same developmental stage or compared to that of GL of a later stages (91 dpc) at p < 0.05 was achieved with considerable low false discovery rates (q ≤ 0.03 and q ≤ 0.14, respectively). Together with the relatively large differences between 63 dpc and 91 dpc in GL and moderate divergence between early and late PI samples as well as between GL 91 dpc and PI 91 dpc this indicates that in GL at 63 dpc a less progressed status has been reached and catching up on the development proceeds until 91 dpc. The assignment of DE-genes to biofunctions and canonical pathways underline the relatively delayed development in GL compared to PI in terms of differentiation, maintenance, and maturation and a thus prolonged proliferative phase.
DE genes essentially belong to signaling pathways
The development of embryonic mammary gland can be divided into several steps as described above. Many known genes were regulated in each stage of mammary development. These genes were assignment to signaling pathways whereas expression of genes of metabolic pathways was not differentially regulated among the samples. Previous studies have already addressed a number of genes of signaling pathways in particular those related to growth factors. Fibroblast growth factor 2 (FGF2) and its receptor (FGFR2) were found differentially expressed in this study (Table 2). Fibroblast growth factor (FGFs) together with T-box transcription factor were found in mammary line in early development and members of the Wnt gene family were also found in mammary placodes. Members of the Wnt signaling pathway were differentially expressed between 63 and 91 dpc. They play a role in the formation of placodes and mammary epithelial differentiation [17-19]. Lymphocyte enhancer factor-1 (LEF1), an important factor for the mammary placodes and bud maintenance, is a transcription factor, which is defined in epithelial cell of mammary buds and mesenchyme [20]. It was found regulated due to breed and stage in this study (Tables 1 and 2). The expression of LEF1 is induced by parathyroid hormone - protein (PTHLP) and its receptor (PTHR1) [21]. PTHLH (parathyroid hormone like hormone) and PTHR1 genes were clearly detected in epithelial and connective teat tissues. Polymorphisms of PTHLH and PTHR1 as well as LEF1 were associated with the inverted teat phenotype [22,23]. Studies of mice deficient of FGFs, Wnts, LEF1 and PTHLH show that these are required for adequate signaling in fetal mammary development [24-28].
The development of fetal mammary gland largely depends on interaction between mesenchymal-epithelial. The basic molecules for signals in mesenchyme-epithelial interaction are provided by paracrine signaling, which consists of epithelial tyrosine kinase receptors and their mesenchymal ligands [29]. Fibroblast growth factor (FGF) signaling that was addressed in this study, like Hepatocyte growth factor (HGF) signaling, Growth hormone signaling, and Epidermal growth factor (EGF) signaling are the main cellular growth, proliferation and development signaling pathways. Accordingly, genes (GAB1, MAPK9, PIK3C2B, PIK3C3 and PRKCH) from signaling pathways were selected to validate microarray data that all have known roles in developmental processes.
GRB2-associated binding protein 1 (GAB1) is a member of the family of docking proteins. GAB1 functions as a mediator in growth factor signaling and cytokine receptor, especially in EGF signaling, HGF signaling and Platelet-derived growth factor (PDGF) signaling which stimulates the migration of epithelial, mesenchymal and hematopoietic cells during embryogenesis and branching morphogenesis of epithelial cells. The deficiency of GAB1 results in the lack of ligands, receptors and signaling molecules and develop the organs [30-34].
Mitogen-activated protein kinase 9 (MAPK9), which is known as c-Jun NH2-terminal kinase 2 (JNK2), is a member of the MAP kinase family. It also enhances in the signal transduction pathways. MAPKs are regulated through cascades composed of MAPK, MAPK kinase (MAPKK, MKK or MEK) and MAPKK kinase or MEK kinase (MAPKKK or MEKK). These kinases are activated through hormones and growth factors that act via RTKs (e.g. EGF, PDGF and FGF) or cytokine receptors (e.g. growth hormone). MAPKs function in gene transcription, protein synthesis, cell cycle, cell death, and cell differentiation. MAPKs may involve in epithelial-mesenchymal transition (EMT). The overexpression of MAPK induced cell migration and invasion, a morphologic change in EMT. The lack of MAPK increasing tumor aneuploidy and reduced DNA damage [35-40].
Phosphoinositide-3-kinase, class 3 (PIK3C3) and phosphoinositide-3-kinase, class 2, beta polypeptide (PIK3C2B) belong to the phophoinositide-3-kinase (PI3K) family. PI3Ks are a family of unique and conserve intracellular lipid kinases that are activated by growth hormones, RTKs and G-protein-coupled receptors (GPCRs). PI3Ks are capable of phosphorylation the 3´-hydroxyl group of phosphatidylinositol 4,5 diphosphate (PIP2) to generate phosphatidylinositol-3,4,5-trisphosphate (PIP3), which activates intracellular signaling pathways for regulated function in cell growth and proliferation, cell migration, and cell metabolism. PI3Ks can induce the differentiation of cells of epithelial or mesenchymal origin. During EMT, TGFß signaling co-operates with Rho signaling to activate PI3K. PRL-3 signals through PI3K and then leads to EMT [41-45]. PIK3C3 homozygous mutant causes cell proliferation defect, embryonic lethal and death [46].
Protein kinase C, eta (PRKCH) is calcium independent and phospholipids dependent member of the protein kinase C (PKC) family. PKC, a family of serine and threonine - specific protein kinases, has important roles in cell proliferation, survival, migration and adhesion. PKC is activated via lipid activators and phosphorylation. PKC are classified into α, ßI, ßII, γ, δ, ϵ, η (L), θ, μ, ξ, λ isoforms. PKC is predominantly a cytosolic enzyme in the mammary gland and have a role in prolactin (PRL) to stimulate lactogenic processes. PKC- α isoform has a role in mammary cell differentiation, while PKC- δ isoform is mediated by MEK/ERK pathway and induce the production of proteolytic enzymes [47-52].
Conclusions
The study provides a holistic view on the gene expression during prenatal development addressing the abundance of 11,731 probe sets at two fetal stages at the phase of growth and protrusion of mammary buds in two pig breeds. It is shown that the development in PI precedes GL in a way that processes of differentiation, maintenance, and maturation are predominant whereas in GL still proliferative processes are prevalent. These differences in fetal development have implications on the postnatal phenotypes. Due to divergent selection pressure, GL and PI differ in the number of teats and in functional morphological criteria of mammary complex quality, like position and symmetry, shape, and functionality. In particular there is large divergence in the prevalence of inverted teats, a disorder characterized by the formation of non-functional teats. We hypothesed that differential expression between the two breeds at fetal stages reflect the prenatal initiation of postnatal phenotypes also in terms of the liability to emerge inverted teats. We have previously identified molecular routes that are differentially regulated in normal compared to inverted teats at peripubertal stages [11,13]. We questioned here, whether these molecular routes are already relevant at prenatal stages and thus their modulation may initiate the postnatal development. In fact, the DE-genes were mainly from signaling pathways that are known to have implication for mammary gland development. The finding of overwhelming importance of signaling pathways in teat development and of differential expression of related genes, which suggest a prolonged proliferative development in the breed GL selected against undesired teat phenotypes and which potentially promotes the emergence of inverted teats, is in line with our previous findings when comparing normal and inverted teats at postnatal stages [13].
Materials and methods
Sample collection and RNA preparation
Animal care and tissue collection processes followed the guidelines of the German Law of Animal Protection, and the experimental protocol was approved by the Animal Care Committee of the Leibniz Institute for Farm Animal Biology (FBN, Dummerstorf, Germany). The milk lines samples were collected from14 fetuses of sows of the breed German Landrace (GL) and the Pietrain (PI) at the 63 and 91 days post conception (dpc) (63 dpc GL n = 4; 91 dpc GL n = 4; 63 dpc PI n = 3; 91 dpc PI n = 3, respectively). After collected, the samples were immediately frozen in liquid nitrogen and stored at −80°C.
For isolation of total RNA, nipples were prepared from the milk line tissues with scalpel and grinded in a mortar under liquid nitrogen. Then they were homogenized with 1 ml Trizol Reagent (Sigma-Aldrich, Taufkirchen, Germany) by using syringes and needles and cleaned up with the NucleoSpin RNA II kit (Macherey-Nagel, Düren, Germany). In addition, the DNAse treatment was done according to the manufacturer’s protocol. After that, the integrity of the RNA samples were checked by visualizing them on 1.5% agarose gel contained formaldehyde stained with ethidium bromide. The concentration levels of RNA were measured by a Nano Drop ND-1000 Spectrophotometer (PEQLAB, Erlangen, Germany). The absence of DNA contamination was checked by using the RNA as a template in PCR amplifying fragments of the glyceraldehydes-3- phosphate dehydrogenase (GAPDH) gene. All RNAs were stored at −80°C for further analysis.
Microarray analysis
Affymetrix GeneChip Porcine Genome Arrays (Affymetrix, St. Clara, USA) containing 24,123 probe sets were used in the expression study. A total 14 RNA samples from different breeds and stages were used for the array hybridization. cDNA was synthesized from total RNA and used to generate biotin-labeled cRNAs target according to the Affymetrix protocol. The biotinylated cRNA was fragmented and used for hybridization to the Affymetrix Gene Chips at 45°C for 16 hours. Hybridization, washing, and scanning of the arrays were done according to the manufacturer’s recommendation. The data were analyzed with the Affymetrix GCOS 1.1.1 software by using the global scaling to a target signal of 500.
Statistical analysis
The raw intensity files (*.cel) were used in microarray data analysis with the Affymetrix Expression Console software (Affymetrix, St. Clara, USA). First the data were processed with the MAS5.0 algorithm to generate probe cell intensity values, i.e. single expression value for each probe set that are derived from intensities of pairs of perfect-match probes and mismatch probes, and to evaluate presence and absence of transcripts. Using default settings (detection p-values of <0.04 for `present´, ≥0.04 and ≤0.06 for `marginal´, and <0.06 for `absent´) only `present´ calls were used. The subsequent data processing, including background correction, probe summarization and normalization, was performed using the probe logarithmic intensity error (PLIER) algorithm that reveal summary values for the probe sets (Affymetrix 2001, 2005). The microarray data related to all samples were deposited in the Gene Expression Omnibus public repository (GEO accession number: GSE32956). The probe intensity value obtained by the PLIER algorithm were transformed to logarithms and evaluated by analysis of variance taking into account the effects of stage and breed and their interaction to detect genes differentially regulated at p < 0.05 (JMP Genomics). Corresponding q-values were calculated according to algorithms established by Storey [53] using QVALUE. Based on BLAST comparison of the Affymetrix porcine target sequences with the porcine genome sequence (Ensembl_Sscrofa_9), 20,689 of the 24,123 probe sets on the Affymetrix Porcine GeneChip were annotated [54]. This source of annotation was used in this study for Ingenuity Pathways Analysis (IPA) (Ingenuity Systems, http://www.ingenuity.com). The Ingenuity applications were used to generate networks and assess statistically relevant biofunctions and canonical pathways associated with the microarray data. The significance of the association between the dataset of differentially expressed genes (DE-genes) and the predefined pathways and functional categories of the Ingenuity Knowledge Base was measured by Fischer’s exact test and adjusted using the Benjamini–Hochberg correction, providing a p-value, which would determine the probability that the association between the genes in the dataset and the pathway is explained by chance alone.
Quantitative real time PCR
1 μg of total RNA of the same individual samples used for microarray analysis were reverse transcribed with Super Script III with Oligo (dT) and random primers (Invitrogen, Karlsruhe, Germany) in a total volume of 10 μl. The cDNAs were used as the template for validation of the gene expression levels by quantitative RT-PCR in duplicate. Quantitative RT-PCR was conducted with the LightCycler 480 system (Roche, Mannheim, Germany). The reaction volume of 10 μl contained 5.0 μl of LightCycler 480 SYBR Green I Master (Roche), 600 nM of each primer, and 2 μl of cDNA. Amplification conditions were 95°C for 10 min, 40 cycles of 95°C for 15 sec, 60°C (annealing) for 10 sec and 72°C for 15 sec. The primers were designed from Affymetrix core sequence with Primer3 (http://frodo.wi.mit.edu/primer3). The list of primer sequences is provided in Table S1 (Additional file 2). Standard curves were derived for each gene from a serial dilution of cDNAs. For all the assays threshold cycles were converted to copy numbers using the standard curves generated by amplifying serial dilutions of an external PCR standard (107 - 102 copies). At the completion of the amplification protocol, all samples were subjected to melting curve analyses and gel electrophoresis to verify the absence of any non-specific product. Copy numbers of the house keeping genes HPRT1 and RPL32 were obtained from each individual sample to enable accounting for variation in RNA input and efficiency of reverse transcription by normalization when calculating mRNA copy numbers of the target genes. Both of the reference genes were checked for lack of variation on the basis of microarray data at prenatal and postnatal stages [13,55]. Data were analyzed like microarray data by analysis of variance including the effects of stage and breed (JMP Genomics). Differences were considered significant at p < 0.05.
Authors’ contributions
Conceived and designed the experiments: SP, EM, KW. Performed the experiments: KC. Analyzed the data: KC, EM, SP, KW. Contributed reagents/materials/analysis tools: SP, EM, KW. Wrote the paper: KC, KW. All authors read and approved the final manuscript.
Supplementary Material
Figure S1. Significant biofunctions (top 20 according to p-value) representing genes differentially expressed between 63 dpc and 91 dpc in Pietrain and German Landrace. All assignments significant after Benjamini–Hochberg correction.
Table S1. List of genes and corresponding primers used for quantitative RT-PCR. 1Reference genes.
Contributor Information
Kunsuda Chomwisarutkun, Email: kunsuda@fbn-dummerstorf.de.
Eduard Murani, Email: murani@fbn-dummerstorf.de.
Siriluck Ponsuksili, Email: s.wimmers@fbn-dummerstorf.de.
Klaus Wimmers, Email: wimmers@fbn-dummerstorf.de.
Acknowledgements
The authors thank the German Federal Ministry of Education and Research (BMBF) and the Förderverein Biotechnologieforschung der Deutschen Schweineproduktion e.V. (FBF) for financial support (FUGATO - HeDiPig, FKZ 0313392A).
References
- Turner CW. The Development of the Mammary Gland. J Anim Sci. 1929;1929:47–51. [Google Scholar]
- Cunha GR, Hom YK. Role of mesenchymal-epithelial interactions in mammary gland development. J Mammary Gland Biol Neoplasia. 1996;1:21–35. doi: 10.1007/BF02096300. [DOI] [PubMed] [Google Scholar]
- Richert MM, Schwertfeger KL, Ryder JW, Anderson SM. An atlas of mouse mammary gland development. J Mammary Gland Biol Neoplasia. 2000;5:227–241. doi: 10.1023/A:1026499523505. [DOI] [PubMed] [Google Scholar]
- Hens JR, Wysolmerski JJ. Key stages of mammary gland development: molecular mechanisms involved in the formation of the embryonic mammary gland. Breast Cancer Res. 2005;7:220–224. doi: 10.1186/bcr1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowin P, Wysolmerski J. Molecular mechanisms guiding embryonic mammary gland development. Cold Spring Harb Perspect Biol. 2010;2:a003251. doi: 10.1101/cshperspect.a003251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson GW, Karpf AB, Kratochwil K. Regulation of mammary gland development by tissue interaction. J Mammary Gland Biol Neoplasia. 1999;4:9–19. doi: 10.1023/A:1018748418447. [DOI] [PubMed] [Google Scholar]
- Hennighausen L, Robinson GW. Signaling pathways in mammary gland development. Dev Cell. 2001;1:467–475. doi: 10.1016/S1534-5807(01)00064-8. [DOI] [PubMed] [Google Scholar]
- Pispa J, Thesleff I. Mechanisms of ectodermal organogenesis. Dev Biol. 2003;262:195–205. doi: 10.1016/S0012-1606(03)00325-7. [DOI] [PubMed] [Google Scholar]
- Borchers N, Reinsch N, Kalm E. Teat number, hairiness and set of ears in a Pietrain cross: variation and effects on performance traits. Arch Tierz. 2002;45:465–480. [Google Scholar]
- Jonas E, Schreinemachers HJ, Kleinwachter T, Un C, Oltmanns I, Tetzlaff S, Jennen D, Tesfaye D, Ponsuksili S, Murani E, Juengst H, Tholen E, Schellander K, Wimmers K. QTL for the heritable inverted teat defect in pigs. Mamm Genome. 2008;19:127–138. doi: 10.1007/s00335-007-9086-5. [DOI] [PubMed] [Google Scholar]
- Tetzlaff S, Murani E, Schellander K, Ponsuksili S, Wimmers K. Differential expression of growth factors and their receptors indicates their involvement in the inverted teat defect in pigs. J Anim Sci. 2009;87:3451–3457. doi: 10.2527/jas.2008-1660. [DOI] [PubMed] [Google Scholar]
- Günther C. Morphologie der sogenannten Stülpzitzen beim Schwein im Vergleich zum histologischen Bild der normalen Zitzen. Freie Universitat, Berlin, Germany; 1984. [Google Scholar]
- Chomwisarutkun K, Murani E, Ponsuksili S, Wimmers K. Microarray analysis reveals genes and functional networks relevant to the predisposition for inverted teats in pigs. J Anim Sci. 2012;90:1–15. doi: 10.2527/jas.2011-4269. [DOI] [PubMed] [Google Scholar]
- Birchmeier C, Birchmeier W. Molecular aspects of mesenchymal-epithelial interactions. Annu Rev Cell Biol. 1993;9:511–540. doi: 10.1146/annurev.cb.09.110193.002455. [DOI] [PubMed] [Google Scholar]
- Cunha GR. Role of mesenchymal-epithelial interactions in normal and abnormal development of the mammary gland and prostate. Cancer. 1994;74:1030–1044. doi: 10.1002/1097-0142(19940801)74:3+<1030::AID-CNCR2820741510>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Cunha GR. Mesenchymal-epithelial interactions: past, present, and future. Differentiation. 2008;76:578–586. doi: 10.1111/j.1432-0436.2008.00290.x. [DOI] [PubMed] [Google Scholar]
- Smalley MJ, Dale TC. Wnt signaling and mammary tumorigenesis. J Mammary Gland Biol Neoplasia. 2001;6:37–52. doi: 10.1023/A:1009564431268. [DOI] [PubMed] [Google Scholar]
- Veltmaat JM, Van VW, Thiery JP, Bellusci S. Identification of the mammary line in mouse by Wnt10b expression. Dev Dyn. 2004;229:349–356. doi: 10.1002/dvdy.10441. [DOI] [PubMed] [Google Scholar]
- Robinson GW. Cooperation of signalling pathways in embryonic mammary gland development. Nat Rev Genet. 2007;8:963–972. doi: 10.1038/nrg2227. [DOI] [PubMed] [Google Scholar]
- Eblaghie MC, Song SJ, Kim JY, Akita K, Tickle C, Jung HS. Interactions between FGF and Wnt signals and Tbx3 gene expression in mammary gland initiation in mouse embryos. J Anat. 2004;205:1–13. doi: 10.1111/j.0021-8782.2004.00309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley J, Dann P, Hong J, Cosgrove J, Dreyer B, Rimm D, Dunbar M, Philbrick W, Wysolmerski J. Parathyroid hormone-related protein maintains mammary epithelial fate and triggers nipple skin differentiation during embryonic breast development. Development. 2001;128:513–525. doi: 10.1242/dev.128.4.513. [DOI] [PubMed] [Google Scholar]
- Tetzlaff S, Chomdej S, Jonas E, Ponsuksili S, Murani E, Phatsara C, Schellander K, Wimmers K. Association of parathyroid hormone-like hormone (PTHLH) and its receptor (PTHR1) with the number of functional and inverted teats in pigs. J Anim Breed Genet. 2009;126:237–241. doi: 10.1111/j.1439-0388.2008.00781.x. [DOI] [PubMed] [Google Scholar]
- Tetzlaff S, Jonas E, Phatsara C, Murani E, Ponsuksili S, Schellander K, Wimmers K. Evidence for association of lymphoid enhancer-binding factor-1 (LEF1) with the number of functional and inverted teats in pigs. Cytogenet Genome Res. 2009;124:139–146. doi: 10.1159/000207521. [DOI] [PubMed] [Google Scholar]
- Chu EY, Hens J, Andl T, Kairo A, Yamaguchi TP, Brisken C, Glick A, Wysolmerski JJ, Millar SE. Canonical WNT signaling promotes mammary placode development and is essential for initiation of mammary gland morphogenesis. Development. 2004;131:4819–4829. doi: 10.1242/dev.01347. [DOI] [PubMed] [Google Scholar]
- Mailleux AA, Spencer-Dene B, Dillon C, Ndiaye D, Savona-Baron C, Itoh N, Kato S, Dickson C, Thiery JP, Bellusci S. Role of FGF10/FGFR2b signaling during mammary gland development in the mouse embryo. Development. 2002;129:53–60. doi: 10.1242/dev.129.1.53. [DOI] [PubMed] [Google Scholar]
- Boras-Granic K, Chang H, Grosschedl R, Hamel PA. Lef1 is required for the transition of Wnt signaling from mesenchymal to epithelial cells in the mouse embryonic mammary gland. Dev Biol. 2006;295:219–231. doi: 10.1016/j.ydbio.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Kronenberg HM, Foley J. Reduced expression of the PTH/PTHrP receptor during development of the mammary gland influences the function of the nipple during lactation. Dev Dyn. 2005;233:794–803. doi: 10.1002/dvdy.20406. [DOI] [PubMed] [Google Scholar]
- Wysolmerski JJ, Philbrick WM, Dunbar ME, Lanske B, Kronenberg H, Broadus AE. Rescue of the parathyroid hormone-related protein knockout mouse demonstrates that parathyroid hormone-related protein is essential for mammary gland development. Development. 1998;125:1285–1294. doi: 10.1242/dev.125.7.1285. [DOI] [PubMed] [Google Scholar]
- Niemann C, Brinkmann V, Birchmeier W. Hepatocyte growth factor and neuregulin in mammary gland cell morphogenesis. Adv Exp Med Biol. 2000;480:9–18. doi: 10.1007/0-306-46832-8_2. [DOI] [PubMed] [Google Scholar]
- Nguyen L, Holgado-Madruga M, Maroun C, Fixman ED, Kamikura D, Fournier T, Charest A, Tremblay ML, Wong AJ, Park M. Association of the multisubstrate docking protein Gab1 with the hepatocyte growth factor receptor requires a functional Grb2 binding site involving tyrosine 1356. J Biol Chem. 1997;272:20811–20819. doi: 10.1074/jbc.272.33.20811. [DOI] [PubMed] [Google Scholar]
- Maroun CR, Holgado-Madruga M, Royal I, Naujokas MA, Fournier TM, Wong AJ, Park M. The Gab1 PH domain is required for localization of Gab1 at sites of cell-cell contact and epithelial morphogenesis downstream from the met receptor tyrosine kinase. Mol Cell Biol. 1999;19:1784–1799. doi: 10.1128/mcb.19.3.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, Hirano T. The role of Gab family scaffolding adapter proteins in the signal transduction of cytokine and growth factor receptors. Cancer Sci. 2003;94:1029–1033. doi: 10.1111/j.1349-7006.2003.tb01396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Yoshida Y, Nishida K, Narimatsu M, Hibi M, Hirano T. Role of Gab1 in heart, placenta, and skin development and growth factor- and cytokine-induced extracellular signal-regulated kinase mitogen-activated protein kinase activation. Mol Cell Biol. 2000;20:3695–3704. doi: 10.1128/MCB.20.10.3695-3704.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felici A, Giubellino A, Bottaro DP. Gab1 mediates hepatocyte growth factor-stimulated mitogenicity and morphogenesis in multipotent myeloid cells. J Cell Biochem. 2010;111:310–321. doi: 10.1002/jcb.22695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb MH. MAP kinase pathways. Prog Biophys Mol Biol. 1999;71:479–500. doi: 10.1016/S0079-6107(98)00056-X. [DOI] [PubMed] [Google Scholar]
- Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev. 2001;81:807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- Chen P, O'Neal JF, Ebelt ND, Cantrell MA, Mitra S, Nasrazadani A, Vandenbroek TL, Heasley LE, Van Den Berg CL. Jnk2 effects on tumor development, genetic instability and replicative stress in an oncogene-driven mouse mammary tumor model. PLoS One. 2010;5:e10443. doi: 10.1371/journal.pone.0010443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Kuiatse I, Lee AV, Pan J, Giuliano A, Cui X. Sustained c-Jun-NH2-kinase activity promotes epithelial-mesenchymal transition, invasion, and survival of breast cancer cells by regulating extracellular signal-regulated kinase activation. Mol Cancer Res. 2010;8:266–277. doi: 10.1158/1541-7786.MCR-09-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Xiong Y, Ma C, Shi T, Ma D. Molecular cloning and characterization of novel human JNK2 (MAPK9) transcript variants that show different stimulation activities on AP-1. BMB Rep. 2010;43:738–743. doi: 10.5483/BMBRep.2010.43.11.738. [DOI] [PubMed] [Google Scholar]
- Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15:740–746. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Wang H, Quah SY, Dong JM, Manser E, Tang JP, Zeng Q. PRL-3 down-regulates PTEN expression and signals through PI3K to promote epithelial-mesenchymal transition. Cancer Res. 2007;67:2922–2926. doi: 10.1158/0008-5472.CAN-06-3598. [DOI] [PubMed] [Google Scholar]
- Kok K, Geering B, Vanhaesebroeck B. Regulation of phosphoinositide 3-kinase expression in health and disease. Trends Biochem Sci. 2009;34:115–127. doi: 10.1016/j.tibs.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Wickenden JA, Watson CJ. Key signalling nodes in mammary gland development and cancer. Signalling downstream of PI3 kinase in mammary epithelium: a play in 3 Akts. Breast Cancer Res. 2010;12:202. doi: 10.1186/bcr2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Takatoh J, Wang F. The mammalian class 3 PI3K (PIK3C3) is required for early embryogenesis and cell proliferation. PLoS One. 2011;6:e16358. doi: 10.1371/journal.pone.0016358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters SB, Rillema JA. Role of protein kinase C in the prolactin-induced responses in mouse mammary gland explants. Mol Cell Endocrinol. 1989;63:159–166. doi: 10.1016/0303-7207(89)90092-0. [DOI] [PubMed] [Google Scholar]
- Kiley SC, Jaken S. Protein kinase C: interactions and consequences. Trends Cell Biol. 1994;4:223–227. doi: 10.1016/0962-8924(94)90146-5. [DOI] [PubMed] [Google Scholar]
- Marte BM, Meyer T, Stabel S, Standke GJ, Jaken S, Fabbro D, Hynes NE. Protein kinase C and mammary cell differentiation: involvement of protein kinase C alpha in the induction of beta-casein expression. Cell Growth Differ. 1994;5:239–247. [PubMed] [Google Scholar]
- Engers R, Mrzyk S, Springer E, Fabbro D, Weissgerber G, Gernharz CD, Gabbert HE. Protein kinase C in human renal cell carcinomas: role in invasion and differential isoenzyme expression. Br J Cancer. 2000;82:1063–1069. doi: 10.1054/bjoc.1999.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossoni VC, Falbo KB, Mauro LV, Krasnapolski MA, Kazanietz MG, Bal De Kier Joffe ED, Urtreger AJ. Protein kinase C delta inhibits the production of proteolytic enzymes in murine mammary cells. Clin Exp Metastasis. 2007;24:513–520. doi: 10.1007/s10585-007-9088-4. [DOI] [PubMed] [Google Scholar]
- Oppenheimer O, Cheung NK, Gerald WL. The RET oncogene is a critical component of transcriptional programs associated with retinoic acid-induced differentiation in neuroblastoma. Mol Cancer Ther. 2007;6:1300–1309. doi: 10.1158/1535-7163.MCT-06-0587. [DOI] [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. J R Stat Soc Ser B. 2002;64:479–498. doi: 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- Naraballobh W, Chomdej S, Murani E, Wimmers K, Ponsuksili S. Annotation and in silico localization of the Affymetrix GeneChip Porcine Genome Array. Arch Tierz. 2010;53:230–238. [Google Scholar]
- Ponsuksili S, Murani E, Phatsara C, Schwerin M, Schellander K, Wimmers K. Porcine muscle sensory attributes associate with major changes in gene networks involving CAPZB, ANKRD1, and CTBP2. Funct Integr Genomics. 2009;9:455–71. doi: 10.1007/s10142-009-0131-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Significant biofunctions (top 20 according to p-value) representing genes differentially expressed between 63 dpc and 91 dpc in Pietrain and German Landrace. All assignments significant after Benjamini–Hochberg correction.
Table S1. List of genes and corresponding primers used for quantitative RT-PCR. 1Reference genes.