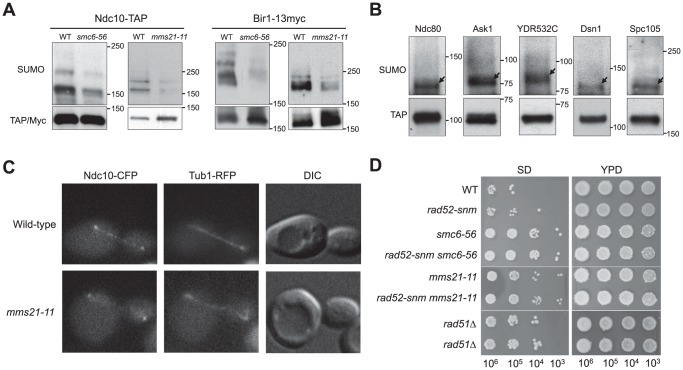
Figure 4. mms21-11 and smc6-56 affect sumoylation of specific kinetochore proteins, Ndc10 localization, and chromosome loss.
(A) Sumoylation of Ndc10 and Bir1 is reduced in mms21-11 and smc6-56 cells. The indicated proteins are tagged at their own chromosomal loci. Sumoylation of these proteins was examined using an anti-SUMO antibody (top panel). The unmodified proteins were detected by anti-TAP or anti-Myc antibody (bottom panel). The slightly different appearance of the SUMO bands is due to different gel percentages. (B) Sumoylation of kinetochore proteins. Sumoylation of each indicated kinetochore protein was examined as in (A). SUMO forms of the proteins are indicated by arrows and migrate at positions approximately 20 kD above the unmodified proteins. (C) Ndc10 spindle localization is defective in mms21-11 cells. Representative anaphase cells containing chromosomally tagged Ndc10-CFP and Tub1-RFP are shown. Note that Ndc10 is found in kinetochores and along spindles in wild-type cells. The spindle localization of Ndc10, but not the kinetochore localization, is defective in mms21-11. (D) mms21-11 and smc6-56 cells exhibit increased loss of chromosomes. Independent isolates of diploid strains were mated with haploid tester strains, and mating products from the indicated number of cells were selected on the SD (synthetic depleted) medium. YPD plates permit the growth of all cells regardless of mating status.