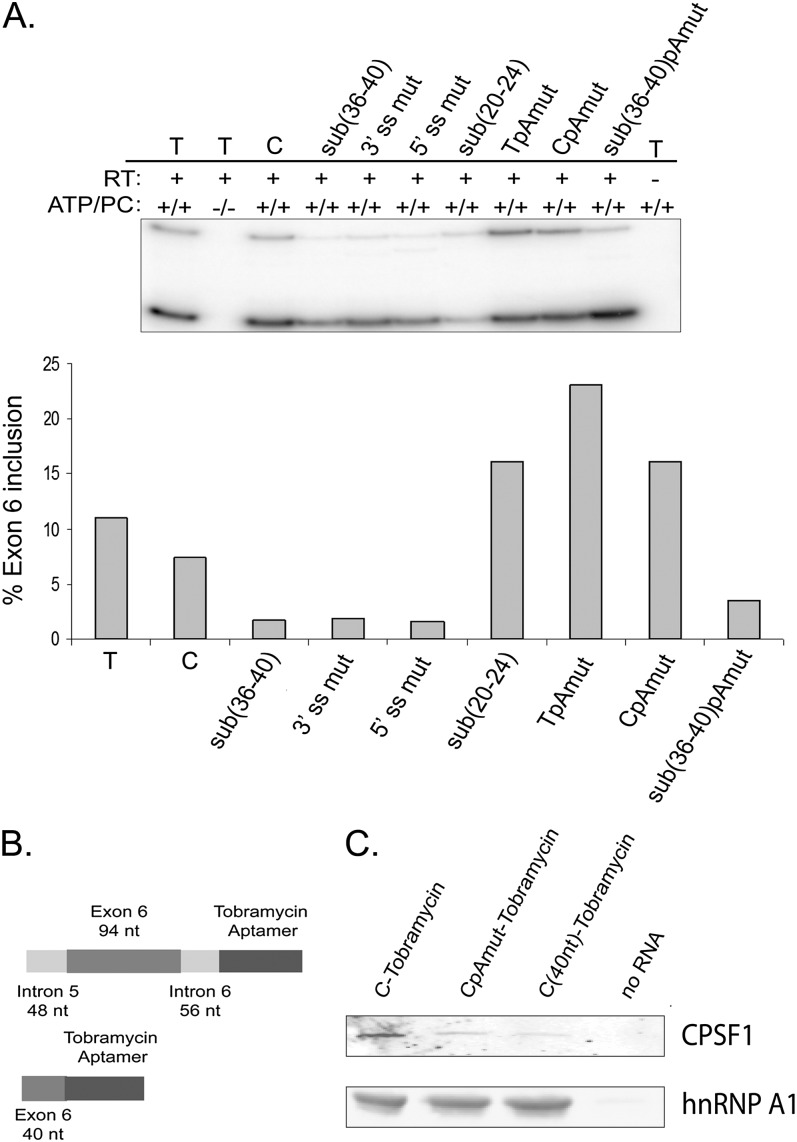
FIGURE 6.
In vitro splicing analysis and RNA affinity chromatography to identify trans-acting factors that regulate splicing of IL7R exon 6. (A) Splicing regulation of IL7R exon 6 is recapitulated in HeLa cell nuclear extracts in vitro. Substitution minigenes described in Figures 1, 2, and 4 were transcribed in vitro, and RNA transcripts used in in vitro splicing reactions with HeLa cell nuclear extracts. “C” and 3′ ss mut and 5′ ss mut minigenes were used as positive and negative controls, respectively. Each bar represents one in vitro splicing reaction. Exon 6 inclusion was analyzed by semiquantitative RT-PCR. This experiment was repeated several times with similar results. (B) RNA transcripts for tobramycin affinity chromatography. (Top) RNAs contained full-length IL7R exon 6 (94 nt), 48, and 56 nt of introns 5 and 6, respectively, followed by tobramycin/streptavidin aptamer sequences. (Bottom) A second set of RNAs contained the first 40 nt of IL7R exon 6, followed by aptamer sequences. Both RNAs were transcribed in vitro from the T7 promoter. (C) Western blot analysis of tobramycin RNA affinity chromatography elution fractions. “C”, CpAmut, and the first 40 nt of exon 6 were utilized for RNA affinity chromatography. Matrix without RNA (“No RNA”) was used as a negative control. The blots were probed with anti-CPSF1 antiserum and an anti-hnRNP A1 antibody.