FIGURE 6.
Illustration of key signal processing steps. (A) Signal decay correction. (Top) An unprocessed full-length electropherogram corresponding to the (+) reagent reaction. (Bottom) The same trace corrected using the algorithm based on Equations 6 and 7. (B) Base calling. Alignment and superimposition of a (−) reagent signal and a γ-scaled (Eq. 8) sequencing signal, both obtained in the same (−) reagent capillary. Aligned peaks in the two signals correspond to individual nucleotides in the RNA sequence. The nucleotide base identity of each peak is indicated by a letter (A, C, G, U). The sequenced base in this experiment was guanosine (G). Note that the G peaks vary in their heights as do the peaks produced by non-G nucleotides, such that it is impossible to unambiguously distinguish G and non-G peaks solely by height. In contrast, the difference in heights of the same peak in the (−) reagent and sequencing signals reliably separates G from non-G peaks. (C) Scaling of (+) reagent and (−) reagent signals. Points correspond to (−) reagent versus (+) reagent termination probabilities, Pterm(i), for 353 nucleotides in the E. coli 16S rRNA obtained in a SHAPE experiment. Nucleotides with the lowest 20% (+) reagent termination probability 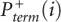 are black; all other nucleotides are blue. The least-squares linear approximation of the black data points is shown as a red line and yields the scaling parameter α (used in Eq. 2).
are black; all other nucleotides are blue. The least-squares linear approximation of the black data points is shown as a red line and yields the scaling parameter α (used in Eq. 2).

