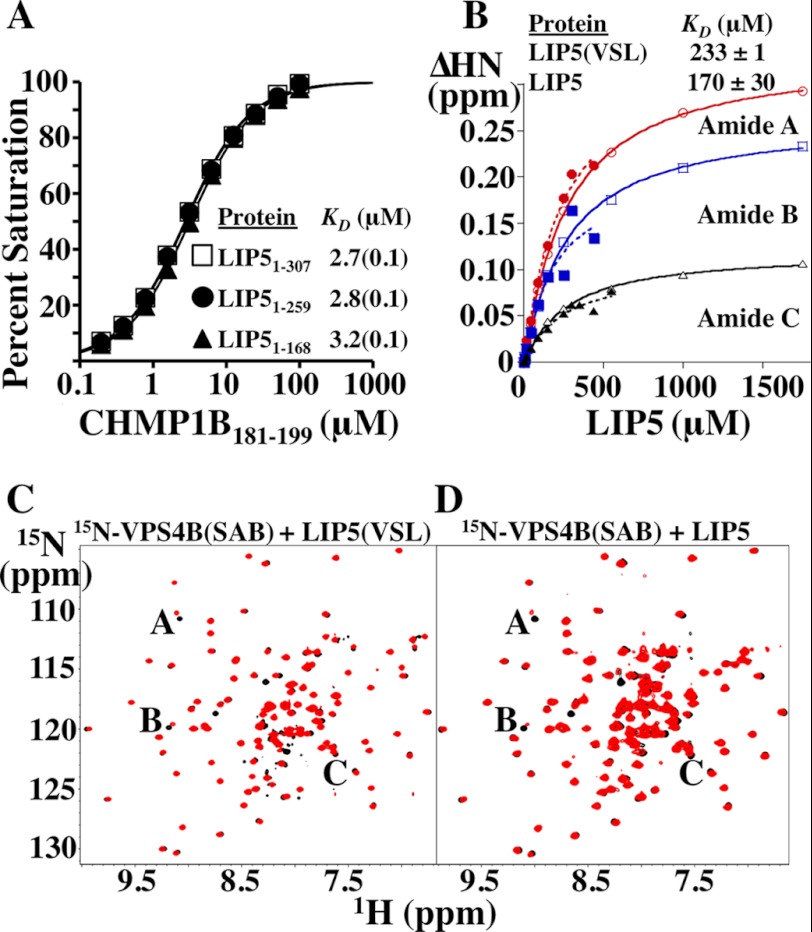
FIGURE 9.
Equilibrium dissociation constants for the LIP5(MIT)2-CHMP1B and LIP5(VSL)-VPS4B complexes are not affected by LIP5 sequence elements outside the minimal binding domains. A, biosensor binding isotherms for CHMP1B(181–199) binding to full-length and C-terminally truncated LIP5 constructs. Data points for each curve were measured in triplicate, and the reported errors in the KD measurements are the estimated errors associated with fitting to simple 1:1 binding models. B, binding isotherms derived from NMR chemical shift perturbations of three different 15N-labeled VPS4B(SAB) backbone amide residues induced by binding of LIP5 (closed symbols, dashed lines) or LIP5(VSL) (open symbols, solid lines). KD values and S.D. reported for LIP5 and LIP5(VSL) binding were from averages of the KD values derived from the individual fits of the 15N-labeled VPS4B(SAB) A, B, and C amide isotherms (see C and D). C, overlaid 1H-15N HSQC spectra of 75 μm 15N-labeled VPS4B(SAB) alone (black) or in the presence of 90 μm LIP5(VSL) (red). Resonances used to derive the binding isotherms shown in B are labeled. D, overlaid 1H-15N TROSY-HSQC spectra of 75 μm 15N-labeled VPS4B(SAB) alone (black) or in the presence of 90 μm full-length LIP5(1–307) (red). Shifted resonances are labeled as described for C. Note that the chemical shifts induced by binding of full-length LIP5 and LIP5(VSL) alone are very similar. Our estimated dissociation constant for the human LIP5(VSL)-VPS4B(SAB) interaction (∼200 μm) is considerably weaker than a previous estimate of the KD for full-length LIP5 binding to immobilized VPS4B/SKD1 (53 nm), but that measurement was made under conditions in which avidity effects could have contributed to the observed KD because the dimeric LIP5 protein was binding immobilized VPS4B/SKD1 (46). Our dissociation constant is ∼3-fold weaker than that reported previously for the yeast Vta1p(VSL)-Vps4p(SAB) interaction (48), and this agreement seems reasonable given that the proteins are from different species and that the previous assay was also configured so that avidity effects could have contributed to the measured binding affinity.