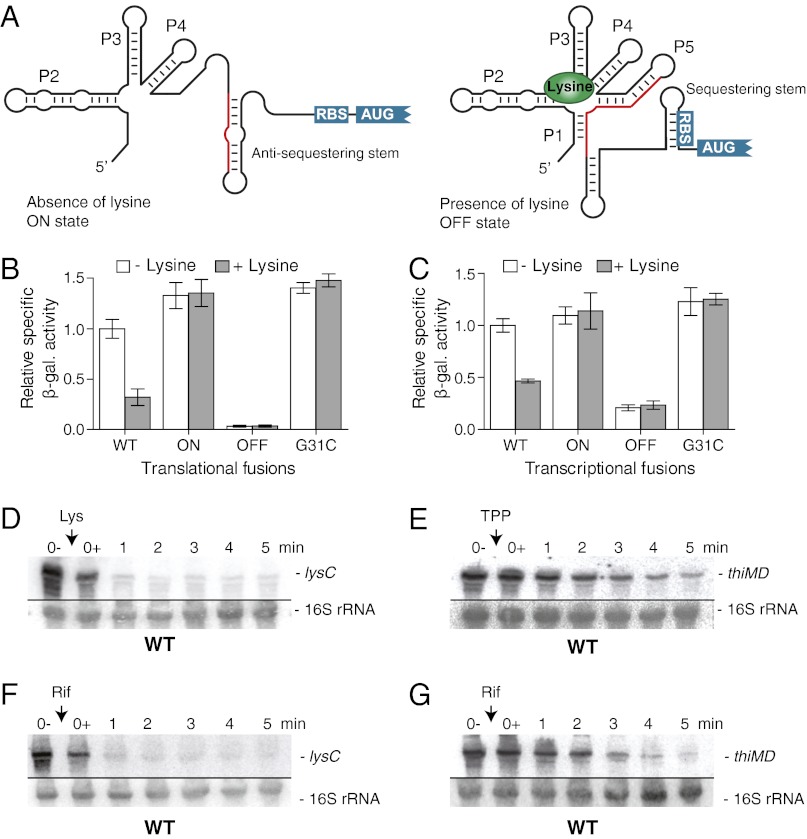
Fig. 1.
The E. coli lysC riboswitch modulates the mRNA level upon lysine binding. (A) Schematic representation of the predicted lysine riboswitch translational control. In the absence of lysine, the ON state exhibits an antisequestering stem, which exposes the RBS and allows translation initiation. However, when bound to lysine, the riboswitch adopts the OFF state in which the presence of a sequestering stem masks the RBS and inhibits gene expression. The region shared by the antisequestering stem (ON state) and the aptamer (OFF state) is represented in red. (B) β-Galactosidase assays of translational LysC-LacZ fusions for wild type and ON, OFF, and G31C mutants (Tables S1–S3). Enzymatic activities were measured in the absence and presence of 10 µg/mL lysine. Values were normalized to the enzyme activity obtained for the wild type in the absence of lysine. (C) β-Galactosidase assays of transcriptional lysC-lacZ fusions for the wild type and for the ON, OFF, and G31C mutants. Enzymatic activities were measured in the absence and presence of 10 µg/mL lysine. Values were normalized to the activity obtained for the wild type in the absence of lysine. (D) Northern blot analysis of lysC mRNA level. Wild-type E. coli strain MG1655 was grown to midlog phase in M63 minimal medium with 0.2% glucose at 37 °C, and total RNA was extracted at the indicated times immediately before (0−) and after (0+) the addition of lysine (10 µg/mL). The probe was designed to detect the riboswitch region (positions 42–201) of the lysC mRNA (SI Appendix, Fig. S4A). 16S rRNA was used as a loading control. (E) Northern blot analysis of the thiMD mRNA level. Bacterial growth culture and RNA extractions were performed as described in D. TPP was added at a final concentration of 500 µg/mL. The probe was designed to detect the riboswitch region (positions 4–141) of thiMD mRNA. 16S rRNA was used as a loading control. (F) Determination of the lysC mRNA stability. Bacterial growth culture and RNA extractions were performed as described in D, but without the addition of lysine. Rifampicin was added at a final concentration of 250 µg/mL. 16S rRNA was used as a loading control. (G) Determination of thiMD mRNA stability. Bacterial growth culture and RNA extractions were performed as described in D. Rifampicin was added at a final concentration of 250 µg/mL. 16S rRNA was used as a loading control.