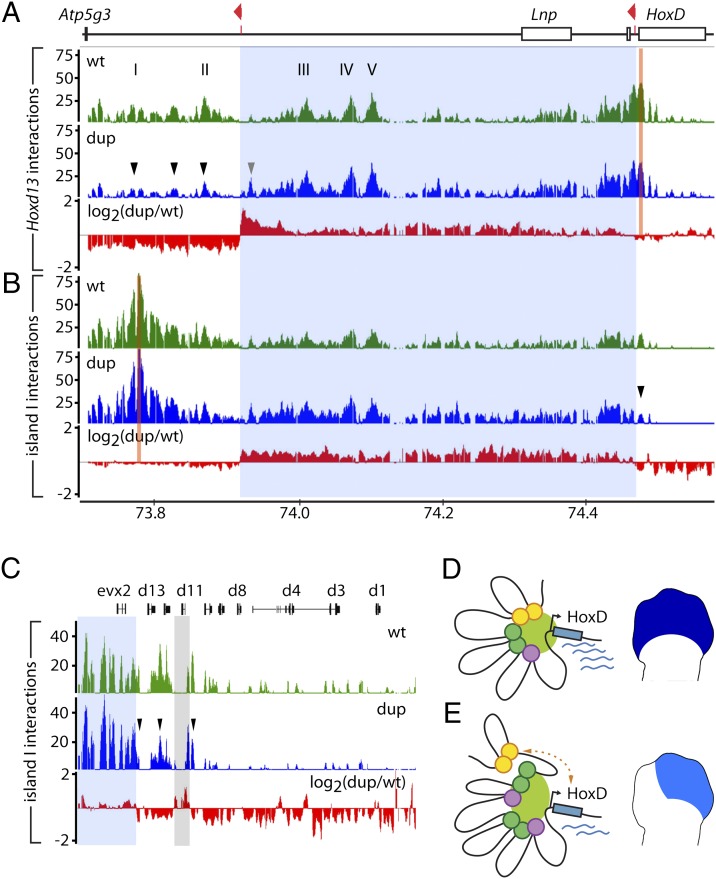
Fig. 7.
The duplication interferes with regulatory interactions. (A) 4C analysis with a viewpoint in Hoxd13 (orange bar) showing long-range interactions over the centromeric gene desert in developing digits of E12.5 WT (green) or Dup(Nsi-SB) (blue) embryos. The red profile displays the ratio (log2 scale) of Dup/WT intensities. The duplicated interval is highlighted in light blue over the profiles. Hoxd13 interactions are increased with sequences within the duplicated segment (gray arrowhead) and decreased with sequences located farther centromeric (black arrowheads). The x axis shows chromosomal coordinates (mm8, 2006 University of California, Santa Cruz assembly) in megabases and the y axes are the ratio of 4C-amplified/genomic DNA intensities. (B) Similar analysis with a viewpoint taken within island I. (C) Enlargement of the HoxD cluster from B. The interactions of island I with the 5′ HoxD cluster are reduced (arrowheads). The light gray bar highlights the sequence corresponding to the Hoxd11LacZ reporter gene present at two positions on this genetic configuration (Fig. 3). (D and E) Distinct conformations in WT or Dup(Nsi-SB) digits, with schematic representation of the transcriptional output in developing digits. (D) In the WT situation, the locus adopts an active conformation, bringing the various islands in the vicinity of the HoxD cluster. (E) The duplication impairs the association of the distal elements (orange circles) with the cluster. The scheme represents one of the possible conformations of the locus, because the interactions experienced by each of the duplicated copies of the islands (purple and green circles) cannot be discriminated with our approach.