Fig. 5.
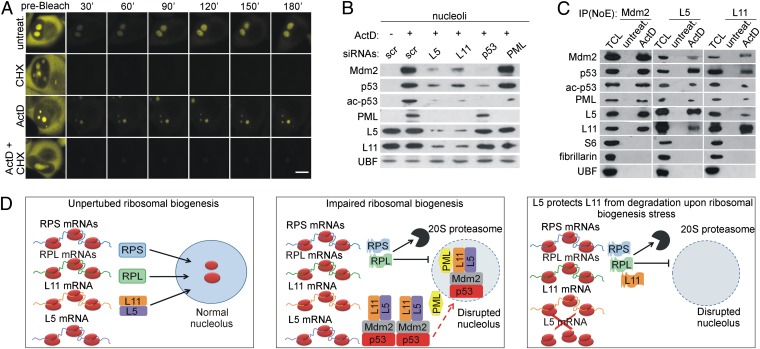
Newly synthesized L5 and L11 colocalize with Mdm2, p53, and PML in the nucleolus upon ribosomal stress. (A) H1299 cells expressing YFP-L11 were treated with ActD for 5 h in the presence or absence of CHX and then photobleached using a 488-nm laser. Time-lapse laser scanning microscopy was used to monitor the recovery of the nucleolar fluorescence signal. (Scale bar, 5 μm.) (B) A549 cells were transfected with siRNAs against L5, L11, p53, or PML for 48 h and then left untreated or treated with ActD for 5 h. Purified nucleoli were immunoblotted with the indicated antibodies. (C) Nucleolar extracts (NoE) from untreated or ActD-treated (5 h) A549 cells were immunoprecipitated with antibodies against Mdm2, L5, or L11. These immunoprecipitates and total cell lysate (TCL) from ActD-treated A549 cells were immunoblotted with the indicated antibodies. (D) (Left) Under normal conditions, newly synthesized RPs of 40S (RPS) and 60S (RPL) ribosomal subunits are imported into the nucleolus. (Middle) Upon impairment of ribosomal biogenesis, the majority of RPL and RPS are synthesized, but they are degraded by nuclear 20S proteasomes. In contrast, L5 and L11 are not degraded, and they accumulate in the nonribosomal fraction, where they bind Mdm2. L5 and L11 colocalize with Mdm2, p53, and PML in the nucleolus after impairment of ribosomal biogenesis, where full p53 activation probably takes place. Less efficient import of newly synthesized L5 and L11 into the nucleolus upon inhibition of ribosomal biogenesis may also contribute to their accumulation in the nonribosomal fraction (indicated by dashed red arrow). (Right) In the absence of L5, ribosome-free L11 is degraded by proteasomes upon ribosomal biogenesis stress, suggesting that L5 and L11 protect each other from degradation and explaining their mutual requirement in p53 activation.