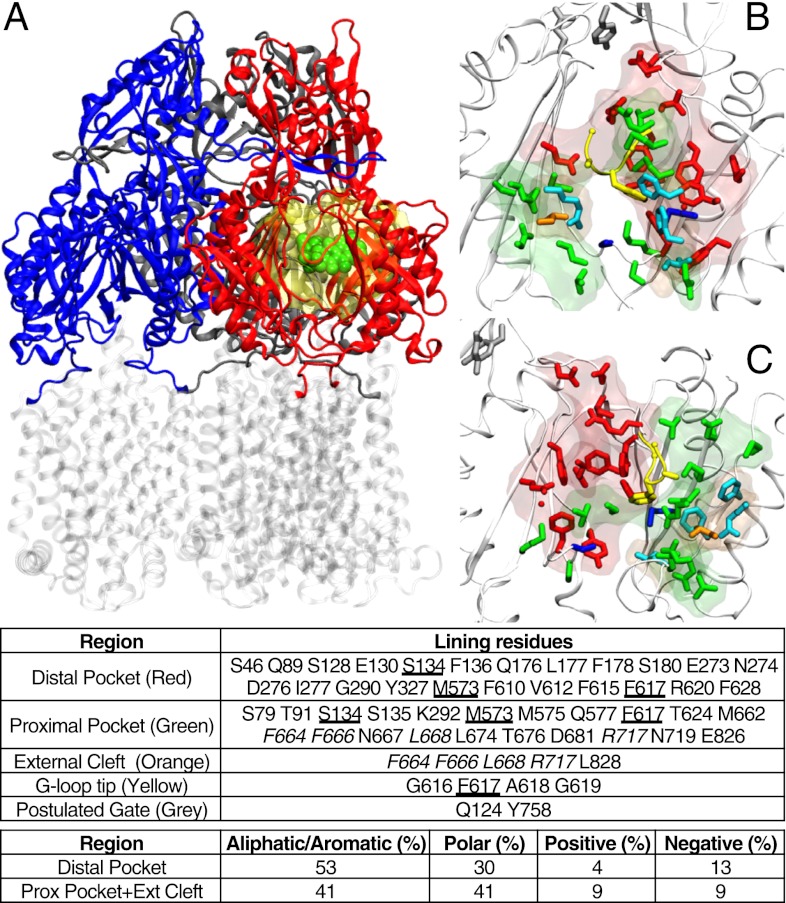
Fig. 1.
(A) Reduced model of AcrB used in this work. The transmembrane domain (in transparent gray) was cut off from the protein, and only the periplasmic domain (blue, red, and gray for access, binding, and extrusion monomer, respectively; residues 33–335 and 565–871 of the intact protein) was kept. The substrate is shown in spheres (green), and the molecular surface of the residues not restrained during the partially restrained MD is shown as a transparent yellow surface. (B and C) Front (B) and side (C) views of the regions of AcrB interacting with substrates (binding monomer; PDB ID code 4DX5). The entrance of the cleft, the proximal, and the distal pockets are shown as transparent surfaces, and the side chains of residues lining them are shown as sticks (orange, green, and red, respectively). Some residues (in addition to those belonging to the G-loop) are shared between the distal and proximal pocket (blue sticks), and four residues of the cleft entrance are also defined as belonging to the proximal cleft (cyan sticks). In addition, the residues of the tip of G-loop and of the gate are shown with yellow and gray sticks respectively. In the table identifying the residues belonging to each region (Lower), those common to the cleft and the proximal pocket are italicized, and those shared between the proximal and the distal pockets are underlined.