Abstract
Background
Yersinia pestis synthesizes the attached biofilms in the flea proventriculus, which is important for the transmission of this pathogen by fleas. The hmsHFRS operons is responsible for the synthesis of exopolysaccharide (the major component of biofilm matrix), which is activated by the signaling molecule 3′, 5′-cyclic diguanylic acid (c-di-GMP) synthesized by the only two diguanylate cyclases HmsT, and YPO0449 (located in a putative operonYPO0450-0448).
Methodology/Principal Findings
The phenotypic assays indicated that the transcriptional regulator Fur inhibited the Y. pestis biofilm production in vitro and on nematode. Two distinct Fur box-like sequences were predicted within the promoter-proximal region of hmsT, suggesting that hmsT might be a direct Fur target. The subsequent primer extension, LacZ fusion, electrophoretic mobility shift, and DNase I footprinting assays disclosed that Fur specifically bound to the hmsT promoter-proximal region for repressing the hmsT transcription. In contrast, Fur had no regulatory effect on hmsHFRS and YPO0450-0448 at the transcriptional level. The detection of intracellular c-di-GMP levels revealed that Fur inhibited the c-di-GMP production.
Conclusions/Significance
Y. pestis Fur inhibits the c-di-GMP production through directly repressing the transcription of hmsT, and thus it acts as a repressor of biofilm formation. Since the relevant genetic contents for fur, hmsT, hmsHFRS, and YPO0450-0448 are extremely conserved between Y. pestis and typical Y. pseudotuberculosis, the above regulatory mechanisms can be applied to Y. pseudotuberculosis.
Introduction
Y. pestis is highly virulent to mammalians including humans, and causes systemic and fatal infections mainly manifested as bubonic, septicemic, and pneumonic plague. Y. pestis is primarily transmitted via the bite of an infected flea. Y. pestis synthesizes the attached biofilms in the flea proventriculus, making the blockage of fleas [1], [2]. The blockage of fleas makes them feel hungry and repeatedly attempt to feed, and thus, the plague bacilli will be pumped into the host body during the futile feeding attempts, promoting the bacterial transmission between mammalian reservoirs [1], [2].
The Yersinia biofilms are a population of bacterial colonies embedded in the self-synthesized extracellular matrix, and the matrix is primarily composed of exopolysaccharide that is the homopolymer of N-acetyl-D-glucosamine [1]. The hmsHFRS operon is responsible for the synthesis and translocation of biofilm exopolysaccharide across the cell envelope, and all the four genes in this operon are required for the biofilm formation and for the flea blockage [1], [3].
The signaling molecule 3′, 5′-cyclic diguanylic acid (c-di-GMP) is a central positive activator of the enzymes catalyzing the production of biofilm exopolysaccharide [4]. HmsT [5], [6] and YPO0449 (y3730 in KIM) [7], [8] are the only two diguanylate cyclase enzymes in Y. pestis to synthesize c-di-GMP, and both of them stimulate the Yersinia biofilm formation. The predominant effect of HmsT was on the in vitro biofilm formation, while the role of YPO0449 in the biofilm production is much greater in the flea than in vitro [7].
The Rcs phosphorelay system negatively controls Yersinia biofilm production in both nematode and flea models [9]. The Rcs system is composed of the sensor kinase RcsC, the phosphotransfer RcsD, and the cytoplasmic response regulator RcsB [10]. RcsC and RcsD transfers phosphate to RcsB, and the phosphorylated RcsB (RcsB-P) binds to some of its target promoters to mediate the gene regulation, whereas a complex of RcsB-P and its accessory protein RcsA is required for the regulation of other target genes [10]. The RcsAB box sequence TAAGAAT-ATTCTTA is a 14 bp inverted repeat [11].
The ferric uptake regulator (Fur) is a predominant iron-regulating system in bacteria [12]. Fur directly controls not only almost of the iron assimilation functions but a variety of genes involved in various non-iron functions, and thus, this regulator governs a complex regulatory cascade in Y. pestis [13], [14]. Two consensus constructs, a 19 bp box and a position-specific scoring matrix (PSSM), have been built to represent the conserved cis-acting signals recognized by Fur [13]. The Fur box sequence AATGATAATNATTATCATT is a 9-1-9 inverted repeat.
During the general maintenance of Y. pestis on the agar media, we found that the fur mutant exhibited a much more rugose and dry colony morphology relative to its parent strain, which promoted us to hypothesize the Fur-mediated repression of exopolysaccharide synthesis and biofilm production in Y. pestis (see below for details). In the present work, the detection of biofilms verified that Fur inhibited the Y. pestis biofilm production in vitro and on nematode. The subsequent gene regulation experiments disclosed that Fur specifically bound to the promoter-proximal region of hmsT for repressing the hmsT transcription, and yet, it had no regulatory effect on hmsHFRS and YPO0450-0448. In addition, the detection of intracellular levels of c-di-GMP revealed that Fur inhibited the c-di-GMP production. Therefore, Y. pestis Fur inhibited the c-di-GMP production through directly repressing the transcription of hmsT, and thus, it acted as a repressor of biofilm formation.
Materials and Methods
Bacterial Strains and Growth
The wild-type (WT) Y. pestis biovar Microtus strain 201 is avirulent to humans but highly lethal to mice [15]. The entire coding region of fur or the base pairs 146 to 468 of hmsS was replaced by the kanamycin resistance cassette by using the one-step inactivation method based on the lambda phage recombination system, to generate the fur or hmsS null mutant (designated as Δfur or ΔhmsS, respectively) of Y. pestis, as described previously [14]. All the primers used in this study were listed in Table 1. Given the pervious observation that the deletion of hmsS lead to a biofilm-defective phenotype in Y. pestis [16], ΔhmsS was used as a reference biofilm-defective strain in this work.
Table 1. Primer used in this study.
| Target gene | Primers (5′-3′; F/R) |
| Mutant construction | |
| fur | CAGCCTTAATTTGAATCGATTGTAACAGGACTGAATCCGCTGTAACGCACTGAGAAGC/GTGCTTAAAATCTTTATAAGAGTAATGCGATAAAACGATAAGATTGCAGCATTACACG |
| hmsS | CGATACCGTTGAGGATTTATATTCTCAGCGGTTTGACGACAGATTGCAGCATTACACG/TATCCAGAACTTTTACGGATTCATAATAGAGTGGCGCGCTTGTAACGCACTGAGAAGC |
| Complementation of the fur mutant | |
| fur | AGACCGCCAACCTGAACTG/CAACGAAGAATAGCCACCTGAC |
| Protein expression | |
| fur | GCGGGATCCATGACTGACAACAACAAAG/GCGAAGCTTTTATCTTTTACTGTGTGCAGA |
| Primer extension | |
| hmsH | /TATTGTTGCAAAGTCATTATAGGAT |
| hmsT | /GGTATTTATTCCGACATCACGAC |
| YPO0450 | /AGTAGCGGTAGTCATTTTTACG |
| LacZ reporter fusion | |
| hmsH | GCGGGATCCACTTTGCTGAAGACTTGTCACG/GCGAAGCTTCCGCCATAGCAGGATTAACG |
| hmsT | GCGGAATTCGCCCAGTACAGGTAACAAGG/GCGGGATCCCTGATCGTAGGAGTGGCTATTC |
| YPO0450 | TCTGGATCCCTTACTGGTTGCTATTGCC/TCTAAGCTTGAGGTTCATGATGTTCATCA |
| EMSA | |
| hmsH | ACTTTGCTGAAGACTTGTCACG/CCGCCATAGCAGGATTAACG |
| hmsT | GCCCAGTACAGGTAACAAGG/CTGATCGTAGGAGTGGCTATTC |
| YPO0450 | CTTACTGGTTGCTATTGCC/GAGGTTCATGATGTTCATCA |
| DNase I footprinting | |
| hmsT | GCCCAGTACAGGTAACAAGG/TTTGTTTCAGCCTGTCATCATG |
| CATGATGACAGGCTGAAACAAA/CTGATCGTAGGAGTGGCTATTC | |
A PCR-generated DNA fragment containing the fur coding region together with its promoter-proximal region (458 bp upstream the coding sequence) and transcriptional terminator (189 bp downstream) was cloned into the pACYC184 vector (GenBank accession number X06403) that harbors a chloramphenicol resistance gene. Upon being verified by DNA sequencing, the recombinant plasmid was introduced into Δfur, yielding the complemented mutant strain C-fur.
The incubation temperature of 26°C was employed for the Y. pestis cultivation, unless otherwise specifically indicated. For the general bacterial cultivation and maintenance, Y. pestis was cultivated in the Luria-Bertani (LB) broth or on the LB agar plate. For preparing the glycerol stocks of bacterial cells, a single colony was inoculated on the LB agar plate for further incubation for 1 to 2 d; the bacterial cells were washed into the LB broth at an optical density at 620 nm (OD620 ) of about 1.5, and stored with addition of 30% glycerol at −80°C. For primer extension or LacZ fusion, 200 µl of bacterial glycerol stocks were inoculated into 18 ml of fresh LB broth, and allowed to grow with shaking at 230 rpm to an OD620 of 0.4 to 0.5 prior to the bacterial harvest.
RNA Isolation and Primer Extension Assay
Before bacterial harvest, double-volume RNAprotect Bacteria Reagent (Qiagen) was added immediately to each cell culture. Total bacterial RNAs were extracted using the TRIzol Reagent (Invitrogen) [17]. RNA quality was monitored by agarose gel electrophoresis, and RNA quantity was determined by spectrophotometry. For the primer extension assay [17], an oligonucleotide primer complementary to a portion of the RNA transcript of each indicated gene was employed to synthesize cDNAs from the RNA templates. One to 10 µg of total RNA from each strain was annealed with 1 pmol of [γ-32P] end-labeled reverse primer using a Primer Extension System (Promega) according to the manufacturer’s instructions. The same labeled primer was also used for sequencing with the fmol® DNA Cycle Sequencing System (Promega). The primer extension products and sequencing materials were concentrated and analyzed in a 6% polyacrylamide/8 M urea gel. The result was detected by autoradiography (Kodak film).
LacZ Reporter Fusion and β-Galactosidase Assay
The promoter-proximal DNA region of each gene tested was prepared by PCR with the Takara ExTaq DNA polymerase by using Y. pestis 201 genome DNA as template, and then cloned directionally into the HindIII-BamHI site of the transcriptional fusion vector pRW50 [18] that contained a promotorless lacZ reporter gene. Correct clone was verified by DNA sequencing. Each Y. pestis strain tested was transformed with the recombinant plasmids. The empty plasmid was also introduced into each strain as negative control. The β-Galactosidase activity was measured on cellular extracts from cells cultivated as above by using the β-Galactosidase Enzyme Assay System (Promega) [17].
Preparation of 6×His-tagged Fur (His-Fur) Protein
To prepare a His-Fur protein [14], the entire coding region of fur was amplified from Y. pestis 201 and cloned directionally into the BamHI and HindIII site of plasmid pET28a (Novagen), which was verified by DNA sequencing. The recombinant plasmids encoding the His-Fur protein were transformed into Escherichia coli BL21 (DE3) cells (Novagen). Expression of His-Fur protein was induced by addition of 1 mM isopropyl-beta-D-thiogalactoside. The His-Fur protein was purified under native conditions with a QIAexpressionist™ Ni-NTA affinity chromatography (Qiagen). The purified, eluted protein was concentrated with the Amicon Ultra-15 (Millipore) to a final concentration of about 0.1 to 0.3 mg/ml in the storage buffer (PBS, pH 7.5 plus 20% glycerol). The protein purity was verified by SDS-PAGE with silver staining. The purified protein was stored at −80°C.
Electrophoretic Mobility Shift Assay (EMSA)
For EMSA [14], promoter-proximal DNA regions were prepared by PCR amplification for EMSA. EMSA was performed using the Gel Shift Assay Systems (Promega). The 5′ ends of DNA were labeled using [γ-32P] ATP and T4 polynucleotide kinase. DNA binding was performed in a 10 µl volume containing binding buffer [100 µM MnCl2, 1 mM MgCl2, 0.5 mM DTT, 50 mM KCl, 10 mM Tris-HCl (pH 7.5), 0.05 mg/ml sheared salmon sperm DNA, 0.05 mg/ml BSA and 4% glycerol], labeled DNA (1000 to 2000 c.p.m/µl) and increasing amounts of His-Fur. We still included two control reactions: one contained the specific DNA competitor (unlabeled promoter DNA regions; cold probe), while the other was the non-specific protein competitor (rabbit anti-F1-protein polyclonal IgG antibody). After incubation at room temperature for 30 min, the products were loaded onto a native 4% (w/v) polyacrylamide gel and electrophoresed in 0.5×TB buffer containing 100 µM MnCl2 for 30 min at 220 V. Radioactive species were detected by autoradiography.
DNase I Footprinting
For DNase I footprinting [14], promoter-proximal DNA regions were prepared by PCR amplification performed with specific primer pairs including a 5′-32P-labeled forward or reverse one and its non-labeled counterpart. The PCR products were purified using Qiaquick columns (Qiagen). Increasing amount of purified His-protein was incubated with the labeled DNA fragment (2 to 5 pmol) for 30 min at room temperature in a final volume of 10 µl containing binding buffer same as EMSA. Before DNA digestion, 10 µl of Ca2+/Mg2+ solution (5 mM CaCl2 and 10 mM MgCl2) was added, followed by incubation for 1 min at room temperature. Then, the optimized RQ1 RNase-Free DNase I (Promega) was added to the reaction mixture, and the mixture was incubated at room temperature for 30 to 90 s. The cleavage reaction was stopped by adding 9 µl of the stop solution (200 mM NaCl, 30 mM EDTA and 1% SDS) followed by DNA extraction and precipitation. The partially digested DNA samples were then analyzed in a 6% polyacrylamide/8 M urea gel. Protected regions were identified by comparison with the sequence ladders. For sequencing, the fmol® DNA Cycle Sequencing System (Promega) was used. The result was detected by autoradiography (Kodak film).
Crystal Violet (CV) Staining of Biofilms
Two-hundred microlitre of bacterial glycerol stocks were spotted on the LB agar plate for further incubation for 1 to 2 d. The resulting bacterial cells were washed into the LB broth with an OD620 value of at least 1.0, stored at 4°C for cold shock for 8 to 12 h, and then diluted to an OD620 value of 0.8 with fresh LB broth. The diluted cultures were transferred into the 24-well tissue culture plates with 1 ml of cultures in each well, and allowed to grow at 230 rpm for 24 h. The media containing the planktonic cells were removed for determining the OD620 values. The well with the adherent biofilms was gently washed three times with 2 ml of H2O, and then incubated at 80°C for 15 min for the fixation of attached cells. The surface-attached cells were stained with 2 ml of 0.1% crystal violet for 15 min. The solution was removed, and the well was washed three times with 2 ml of H2O. Bound dye in the well was dissolved with 3 ml of dimethylsulfoxide. The OD570 values were recorded to indicate the crystal violet staining. The OD570/OD620 values were calculated to indicate the relative biofilm formation. The OD620 values were used for normalization to avoid the effect of growth rate and cell density.
Caenorhabditis Elegans Biofilm Assays
The lawns of biofilm-negative Escherichia coli OP50, a uracil auxotroph whose growth was limited on the NGM (Nematode Growth Medium) agar plates, were used as the standard foods for C. elegans. When the larvae or adults of C. elegans grow on the lawns of Y. pestis, this bacterium creates biofilms to cover primarily on the nematode head by blanketing the mouth and thus inhibiting the nematode feeding, which has been developed as a model for Yersinia biofilm research [19], [20]. Bacterial strains were transformed with the pBC-GFP vector [21] to generate Y. pestis WT-GFP, Δfur-GFP, ΔhmsS-GFP, and E. coli OP50-GFP, respectively. To make the bacterial lawns, 200 µl of bacterial glycerol stocks were spotted on the LB agar plate for further incubation for 1 to 2 d. The resulting bacterial cells were washed into the LB broth with an OD620 value of at least 1.5, and aliquots of 300 µl were spotted on the LB agar plate for further incubation for 24 h. About 30 nematodes (adults or L4-stage larvae) were placed on each bacterial lawn expressing GFP, followed by incubation at 20°C for 12 h. The nematodes were suspended in the sterile M9 buffer (4.2 mM Na2HPO4, 2.2 mM KH2PO4, 8.55 mM NaCl, and 1 mM MgSO4) and then washed twice with M9 to remove planktonic bacteria. Worms was examined immediately by the epifluorescence microscopy.
Colony Morphology Assay
Aliquots of 5 µl of bacterial glycerol stocks were spotted on the LB plate, followed by the incubation for one week. The photograph of surface morphology of each bacterial colony was recorded.
Determination of Intracellular Levels of c-di-GMP
The intracellular levels of c-di-GMP were determined by a chromatography-coupled tandem mass spectrometry (HPLC-MS/MS) method as described previously [22]. Two-hundred microlitre of bacterial glycerol stocks were spotted on the LB agar plate for further incubation for 1 to 2 d. The resulting bacterial cells were washed into the LB broth with an OD620 value of about 0.5, and then aliquots of 5 ml were harvested for the extraction of c-di-GMP with the extraction solvent (acetonitrile: methanol:water = 40∶40∶20, v/v/v). The chromatographic separation was performed on a Spark HPLC system equipped with a binary pump system and a 200 µl sample loop. The analyte detection was performed on an API 4000-QTRAP quadrupole mass spectrometer equipped with an electro spray ionization source (Applied Biosystems).The serially diluted water solutions of HPLC-grade c-di-GMP (KeraFAST) were used for determining the standard curves. The HPLC-grade xanthosine 3′,5′-cyclic monophosphate (c-XMP, Sigma) was added as the internal standard into the c-di-GMP extract or standard solution at a final concentration of 50 ng/ml. Aliquots of 1 ml of bacterial cultures were harvest, and the amount of whole-cell protein was determined with a Micro BCA Protein Assay Kit (Thermo Scientific). The final c-di-GMP concentrations were expressed as pmol/mg of bacterial protein.
Experimental Replicates and Statistical Methods
For phenotypic assays and LacZ fusion, experiments were performed with at least three independent bacterial cultures, and the values were expressed as mean ± standard deviation. Paired Student’s t-test was performed to determine statistically significant differences, and P<0.01 was considered to indicate statistical significance. For primer extension, EMSA, and DNase I footprinting, the representative data from at least two independent biological replicates were shown.
Computational Promoter Analysis
The 300 bp upstream regions of the genes tested (Table 2) were retrieved with the ‘retrieve-seq’ program [23]. The PSSM [13] representing the conserved signals for Fur recognition in Y. pestis was used for the pattern matching within the target upstream DNA regions, by using the matrices-paster tool [23].
Table 2. Computational promoter analysis.
| Operon | First gene | Patten matching | ||
| Fur box-like sequence | Position δ | Sore | ||
| hmsT | hmsT | AATGATAATCATAACCAAT | D-272…−254 | 15.07 |
| AACAATAATAATTCCCAAC | D-95…−77 | 8.74 | ||
| hmsHFRS | hmsH | AATGATGATGAAATGGAAT | R-94…−76 | 4.58 |
| YPO0450-0448 | YPO0450 | AATAAGATTTAAGATAAAT | D-139…−121 | 3.89 |
A PSSM [13] representing the conserved signals for Fur recognition in Y. pestis was used for the prediction of Fur-box like sequences within the 300 bp upstream DNA regions of the major biofilm-required genetic loci hmsT, hmsHFRS, and YPO0450-0498. The diguanylate cyclase gene YPO0449 was located in the putative operon YPO0450-0498. δ, the minus numbers indicated the nucleotide positions upstream of translation start, and D and R represented the direct and reverse sequences, respectively.
Results
Fur Inhibited Biofilm Formation
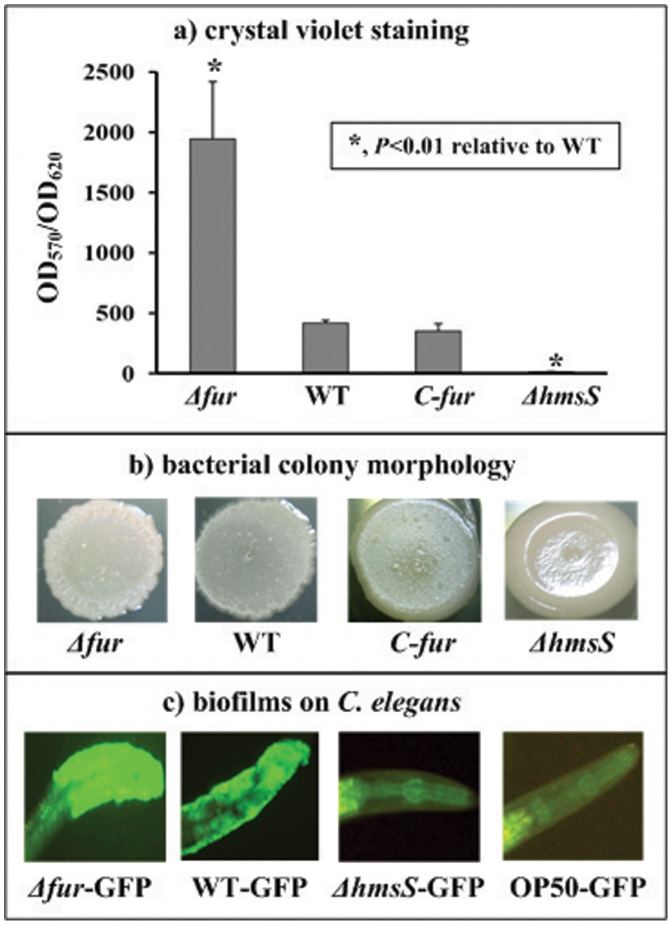
Growing in the polystyrene microtiter plate, Y. pestis cells tend to attach to the walls [9]. The attached biomass (i.e., in vitro biofilms) can be detected with CV staining, which has been developed long time ago as a model for the determination of in vitro biofilms [24]. Herein, Δfur gave the normalized CV staining significantly greater than WT that was comparable to the complemented mutant C-fur, while the biofilm-negative strain ΔhmsS gave almost no CV straining (Fig. 1a).
Figure 1. Yersinia pestis biofilms assays.
a) Adherent bacterial biomass determined by crystal violet staining. Y. pestis was grown in the 24-well polystyrene dishes, and the biomass adherent to the well wall was stained with crystal violet to determine the OD570 values. The planktonic cells were subjective to determine the OD620 values (i.e., cell density) for normalization. Shown were the OD570/OD620 values representing the relative capacity of biofilm formation of each strain tested. b) Bacterial colony morphology. Aliquots of 5 µl of bacterial glycerol stocks were spotted on the LB plate, followed by the incubation for one week. c) Yersinia biofilms on C. elegans . The adult or L4 nematodes were spread on the lawn of Y. pestis expressing GFP and allowed to grow for 12 h. Shown were biofilms attach to the head posterior to the nematode mouth.
Biofilm-forming bacteria growing on the agar plate can give a rugose colony morphology in which the cells are embedded in abundant biofilm exopolysaccharide, and the degrees of rugose colony morphology positively reflect the ability to synthesize the biofilm exopolysaccharide [9], [25], [26]. Δfur produced colonies with much more rugose morphology in relative to WT that was comparable to C-fur, while ΔhmsS made the smooth colonies (Fig. 1b). These suggested that Δfur overproduced the biofilm exopolysaccharide relative to WT.
Yersinia biofilms adhere to the surface of C. elegans, primarily on the head to cover the mouth. When the adult or L4 nematodes were placed on the lawn of Y. pestis expressing GFP and allowed to grow for 12 h, Δfur-GFP produced more extensive and denser biofilms than WT-GFP, while no biofilm was detectable for ΔhmsS-GFP (negative control) and E. coli OP50 (blank control) (Fig. 1c).
Taken together, Y. pestis Fur acted as a repressor for the biofilm formation, most likely through inhibiting the production of biofilm exopolysaccharide.
hmsT was Predicted to be a Direct Fur Target
The Fur PSSM [13] was used to statistically predict the presence of Fur box-like elements [14] within the promoter-proximal regions of the three major biofilm-required loci hmsHFRS, hmsT, and YPO0450-0448. This analysis generated a weight score for each target promoter, and the higher score value indicated the higher probability of the Fur-promoter association [14]. When a frequently used score of 7 was taken as the cutoff value, Fur box-like sequences were found for hmsT rather than the remaining two (Table 2). This computational promoter analysis suggested that Fur could recognize the hmsT promoter for transcriptional regulation.
Fur Repressed hmsT Transcription in a Direct Manner
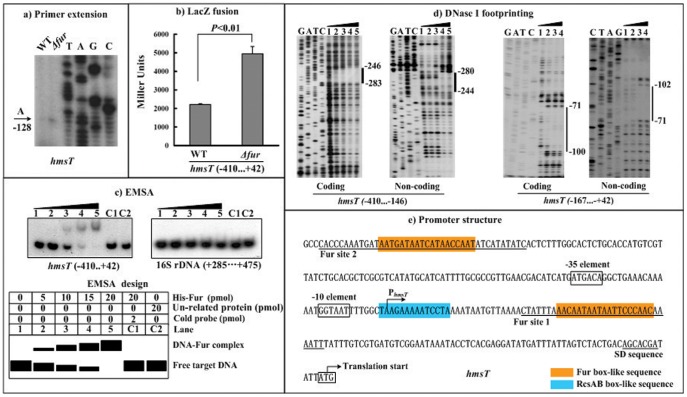
The primer extension experiments (Fig. 2a) were conducted to determine the yield of primer extension product of hmsT (i.e., the relative hmsT transcription level) in WT or Δfur. A single transcriptional start site was detected to be located at the nucleotide A that was 128 bp upstream of hmsT, and thus, a single promoter was transcribed for hmsT. The primer extension assay also disclosed that the mRNA level of hmsT considerably enhanced in Δfur relative to WT.
Figure 2. Repression of hmsT by Fur.
The positive and minus numbers indicated the nucleotide positions upstream and downstream of the translation start, respectively. Lanes G, A, T and C represented the Sanger sequencing reactions. a) Primer extension. An oligonucleotide primer was designed to be complementary to the RNA transcript of hmsT. The primer extension products were analyzed with 8 M urea-6% acrylamide sequencing gel. Shown with the arrow was the transcription start of hmsT. b) LacZ fusion. A promoter-proximal region of hmsT was cloned into the lacZ transcriptional fusion vector pRW50, and transformed into WT or Δfur to determine the hmsT promoter activity, i.e., the β-Galactosidase activity (Miller units) in the cellular extracts. c) EMSA. The radioactively labeled promoter-proximal DNA fragment of hmsT was incubated with increasing amounts of purified His-Fur protein, and then subjected to 4% (w/v) polyacrylamide gel electrophoresis; with the increasing amounts of His-Fur, the band of free target DNA disappeared, and a retarded DNA band with decreased mobility turned up, which presumably represented the protein-DNA complex. A DNA fragment from the coding region of the 16S rRNA gene served as a negative control. d) DNase I footprinting. The labeled coding or non-coding DNA probes were incubated with various amounts of purified His-Fur (lanes 1, 2, 3 and 4, and 5 contained 0, 5, 10, 15 and 20 pmol, respectively), and subjected to DNase I footprinting assay. The protected regions (bold line) were indicated on the right-hand side. e) Promoter structure. Shown were translation/transcription starts, SD sequences, promoter −10 and −35 elements, Fur sites, and Fur/RcsAB box-like sequences for hmsT.
To test the action of Fur on the promoter activity of hmsT, we constructed an hmsT::lacZ fusion vector, containing a 453 bp promoter-proximal region of hmsT and the promoterless lacZ, which was then transformed into WT or Δfur (Fig. 2b). The β-galactosidase activity was measured for evaluating the hmsT promoter activity in each strain. The LacZ fusion experiments disclosed that the hmsT promoter activity significantly enhanced in Δfur relative to WT.
EMSA was conducted to answer whether Fur would bind to the hmsT upstream region in vitro (Fig. 2c). As expected, a purified His-Fur bound to the labeled hmsT promoter DNA in a dose-dependent manner. To confirm the specificity of Fur-DNA association, the EMSA experiments still included a partial coding region of the 16S rRNA gene, and the negative results were obtained.
In order to locate the precise Fur sites, DNase I footprinting experiments were performed with both coding and non-coding strands of target DNA fragments (Fig. 2d). Since two Fur box-like sequences were predicted for hmsT, two distinct hmsT promoter-proximal regions, containing the above predicted elements respectively, were subjected to the footprinting experiments. The results confirmed the binding of His-Fur to the two target DNA fragments in vitro. His-Fur protected a single region within each of the two target DNA fragments tested against DNase I digestion in a dose-dependent pattern. The two footprints were located from 283 to 244 bp (site 2) and from 102 to 71 bp (site 1) upstream of hmsT, respectively. Both of them contained the Fur box-like sequences, and were considered as the Fur sites for hmsT (Fig. 2e).
Fur had no Regulatory Effect on hmsHFRS and YPO0450-0448
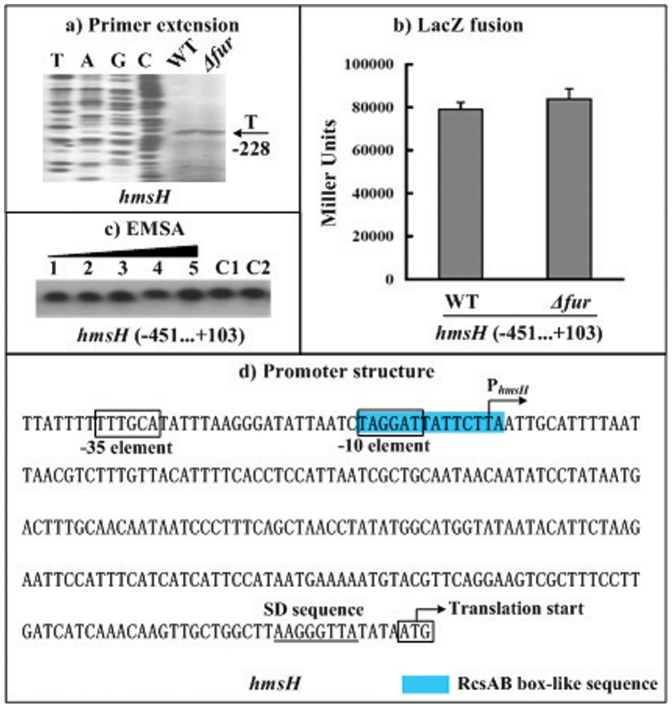
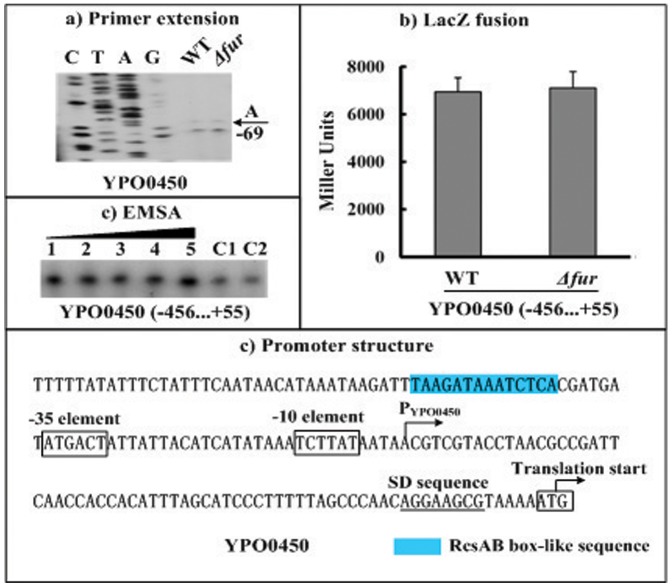
The gene regulation experiments still included the first genes (hmsH and YPO0450) of the hmsHFRS and YPO0450-0448 operons. The primer extension (Fig. 3a and 4a) and LacZ fusion (Fig. 3b and 4b) assays were conducted for hmsH and YPO0450. It was revealed that the fur null mutation have no influence on the hmsH/YPO0450 transcription (Fig. 3a and 4a) or on the hmsH/YPO0450 promoter activity (Fig. 3b and 4b). In addition, the EMSA experiments (Fig. 3c and 4c) indicated that His-Fur could not bind to the upsteam DNA regions of hmsH and YPO0450. Therefore, the Fur regulator had no regulatory action on hmsHFRS and YPO0450-0448 at the transcriptional level under the growth conditions tested herein.
Figure 3. Fur had no regulatory action on hmsH.
The positive and minus numbers of position indicated the nucleotide positions upstream and downstream of the translation start, respectively. a) Primer extension. An oligonucleotide primer was designed to be complementary to the RNA transcript of hmsH. The primer extension products were analyzed with 8 M urea-6% acrylamide sequencing gel. Lanes C, T, A, and G represented the Sanger sequencing reactions. Shown with the arrow was the transcription start of hmsH. b) LacZ fusion. A promoter-proximal region of hmsH was cloned into the lacZ transcriptional fusion vector pRW50, and transformed into WT or Δfur to determine the hmsH promoter activity (Miller units) in the cellular extracts. e) Promoter structure. Shown were translation/transcription starts, SD sequences, promoter −10 and −35 elements, and RcsAB box-like sequence for hmsH.
Figure 4. Fur had no regulatory action on YPO0450.
The positive and minus numbers of position indicated the nucleotide positions upstream and downstream of the translation start, respectively. a) Primer extension. An oligonucleotide primer was designed to be complementary to the RNA transcript of YPO0450. The primer extension products were analyzed with 8 M urea-6% acrylamide sequencing gel. Lanes C, T, A, and G represented the Sanger sequencing reactions. Two closely neighboring extension products were detected. Only the longer product was chosen for identifying the transcription start site shown with the arrow, due to the facts that the shorter extension product might represent the premature stops resulted from the difficulty of polymerase in passing difficult nucleotide sites, and that the core promoter −35 element could not be predicted for the shorter extension product. b) LacZ fusion. A promoter-proximal region of YPO0450 was cloned into the lacZ transcriptional fusion vector pRW50, and transformed into WT or Δfur to determine the YPO0450 promoter activity (Miller units) in the cellular extracts. e) Promoter structure. Shown were translation/transcription starts, SD sequences, promoter −10 and −35 elements, and RcsAB box-like sequence for YPO0450.
Fur Repressed c-di-GMP Production
The intracellular levels of c-di-GMP were determined in WT and Δfur by a HPL-MC/MS method. Compared to WT, a significantly enhanced production of c-di-GMP was observed for Δfur (Fig. 5). These results verified that the Fur-mediated transcriptional repression hmsT accounted for the inhibition of c-di-GMP synthesis by Fur in Y. pestis.
Figure 5. Production of c-di-GMP in different strains.
The intracellular c-di-GMP concentrations were determined by a HPLC-MS/MS method, and the determining values were expressed as pmol/mg of bacterial protein (see supplementary Fig. S1 for representative HPLC-MS/MS traces).
Discussion
Y. pestis is a recently (from the evolutionary point of view) merged clone of the mild enteric pathogen Y. pseudotuberculosis [27]. Y. pseudotuberculosis is transmitted by the food-borne route, while Y. pestis utilizes a radically different mechanism of transmission that rely primarily upon bite of fleas [28]. All of the known structural genes required for the biofilm formation are harbored in Y. pseudotuberculosis, but typical Y. pseudotuberculosis cannot synthesize adhesive biofilms on nematodes and make blockage in fleas [29].
The Y. pseudotuberculosis NghA is a glycosyl hydrolase that cleaves the β-linked N-acetylglucosamine residues, and thus, it plays a key role in degrading the biofilm exopolysaccharide [30].
The RcsAB box-like sequence can be predicted within the promoter-proximal regions of hmsT (Fig. 2e), hmsHFRS (Fig. 3e), and YPO0450-0448 (Fig. 4e). Repression of the hmsT transcription by RcsAB through the RcsAB-promoter association has been established recently [31]. hmsHFRS and YPO0450-044 appears to be the additional direct RcsAB targets (unpublished data), and thus, RcsAB acts as a repressor of Yersinia biofilm formation through inhibiting the production of both c-di-GMP and biofilm exopolysaccharide.
Data presented here disclosed that the Fur regulator had a negative effect on the biofilm formation through repressing the hmsT transcription. DNase I footprinting experiments precisely determined the Fur sites for hmsT. The primer extension assays mapped a single promoter transcribed for hmsT, and accordingly, the core promoter −10 and −35 elements for RNA polymerase recognition were predicted. Collection of data on the translation/transcription start sites, Shine-Dalgarno (SD) sequence (a ribosomal binding site in the mRNA), core promoter −10 and −35 elements for RNA polymerase recognition, and two cis-acting sites for Fur recognition, enabled us to depict the organization of Fur-dependent promoter of hmsT herein (Fig. 2e).
The two Fur sites were located downstream and upstream of the transcription start site of hmsT, respectively, while the RcsAB box-like sequence overlapped the hmsT transcription start. The binding of Fur or RcsAB to the hmsT promoter regions would block the entry of the RNA polymerase to repress the hmsT transcription. In addition, no change in the transcription of hmsHFRS or YPO0450-0448 was observed in the fur mutant compared to its parent strain, indicating that Fur had no regulatory activity on hmsHFRS and YPO0450-0448.
Since the genomic regions encoding Fur, HmsT, HmsHFRS, and YPO0450-0448 were extremely conserved between Y. pestis and typical Y. pseudotuberculosis [32], the regulatory circuit determined herein could be applied to Y. pseudotuberculosis. The action of at least three anti-biofilm factors NghA, RcsAB, and Fur will bring a tight biofilm-negative phenotype of typical Y. pseudotuberculosis. In contrast, Y. pestis has undergone the evolution of loss-of-function of NghA [33] and RcsA [9], which will confer a selective advantage to the progenitor Y. pestis. The mutational loss of function of Fur is of virtual impossibility, since Fur is a predominant regulator of iron assimilation in Y. pestis [13], [14]. Fur-mediated repression of hmsT expression and c-di-GMP synthesis would greatly contribute to finely modulate Yesinia biofilm production within the physiological range. Moreover, Y. pestis has acquired an additional factor Ymt that promotes the bacterial survival of in fleas [30]. The above evolutionary events make Y. pestis prerequisitely survive in fleas and moreover synthesize adhesive biofilms in flea proventriculus to make the blockage, resulting in an efficient arthropod-borne transmission [34].
Supporting Information
Representative HPLC-MS/MS traces. c-di-GMP in a extract of WT (a) or Δfur (b), and a standard sample of c-di-GMP (c) in water at a concentration of 0.3 nM were detected by HPLC-MS/MS -di-GMP c-di-GMP. cXMP was used as the internal standard at a concentration of 50 ng/ml.
(JPG)
Funding Statement
Financial support was provided by the National Natural Science Foundation of China (30930001, and 30900823), by the National Basic Research Program of China (2009CB522600), and by the Beijing Nova Program (Z12111000250000). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hinnebusch BJ, Erickson DL (2008) Yersinia pestis biofilm in the flea vector and its role in the transmission of plague. Curr Top Microbiol Immunol 322: 229–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Darby C (2008) Uniquely insidious: Yersinia pestis biofilms. Trends Microbiol 16: 158–164. [DOI] [PubMed] [Google Scholar]
- 3. Bobrov AG, Kirillina O, Forman S, Mack D, Perry RD (2008) Insights into Yersinia pestis biofilm development: topology and co-interaction of Hms inner membrane proteins involved in exopolysaccharide production. Environ Microbiol 10: 1419–1432. [DOI] [PubMed] [Google Scholar]
- 4. Cotter PA, Stibitz S (2007) c-di-GMP-mediated regulation of virulence and biofilm formation. Curr Opin Microbiol 10: 17–23. [DOI] [PubMed] [Google Scholar]
- 5. Kirillina O, Fetherston JD, Bobrov AG, Abney J, Perry RD (2004) HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis . Mol Microbiol 54: 75–88. [DOI] [PubMed] [Google Scholar]
- 6. Simm R, Fetherston JD, Kader A, Romling U, Perry RD (2005) Phenotypic convergence mediated by GGDEF-domain-containing proteins. J Bacteriol 187: 6816–6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sun YC, Koumoutsi A, Jarrett C, Lawrence K, Gherardini FC, et al. (2011) Differential control of Yersinia pestis biofilm formation in Vitro and in the flea vector by two c-di-GMP diguanylate cyclases. PLoS One 6: e19267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bobrov AG, Kirillina O, Ryjenkov DA, Waters CM, Price PA, et al. (2011) Systematic analysis of cyclic di-GMP signalling enzymes and their role in biofilm formation and virulence in Yersinia pestis . Mol Microbiol 79: 533–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sun YC, Hinnebusch BJ, Darby C (2008) Experimental evidence for negative selection in the evolution of a Yersinia pestis pseudogene. Proc Natl Acad Sci U S A 105: 8097–8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Majdalani N, Gottesman S (2005) The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol 59: 379–405. [DOI] [PubMed] [Google Scholar]
- 11. Wehland M, Bernhard F (2000) The RcsAB box. Characterization of a new operator essential for the regulation of exopolysaccharide biosynthesis in enteric bacteria. J Biol Chem 275: 7013–7020. [DOI] [PubMed] [Google Scholar]
- 12. Escolar L, Perez-Martin J, de Lorenzo V (1999) Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol 181: 6223–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou D, Qin L, Han Y, Qiu J, Chen Z, et al. (2006) Global analysis of iron assimilation and fur regulation in Yersinia pestis. FEMS Microbiol Lett 258: 9–17. [DOI] [PubMed] [Google Scholar]
- 14. Gao H, Zhou D, Li Y, Guo Z, Han Y, et al. (2008) The iron-responsive Fur regulon in Yersinia pestis . J Bacteriol 190: 3063–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou D, Tong Z, Song Y, Han Y, Pei D, et al. (2004) Genetics of metabolic variations between Yersinia pestis biovars and the proposal of a new biovar, microtus. J Bacteriol 186: 5147–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Forman S, Bobrov AG, Kirillina O, Craig SK, Abney J, et al. (2006) Identification of critical amino acid residues in the plague biofilm Hms proteins. Microbiology 152: 3399–3410. [DOI] [PubMed] [Google Scholar]
- 17. Zhang Y, Gao H, Wang L, Xiao X, Tan Y, et al. (2011) Molecular characterization of transcriptional regulation of rovA by PhoP and RovA in Yersinia pestis . PLoS One 6: e25484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. El-Robh MS, Busby SJ (2002) The Escherichia coli cAMP receptor protein bound at a single target can activate transcription initiation at divergent promoters: a systematic study that exploits new promoter probe plasmids. Biochem J 368: 835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Darby C, Hsu JW, Ghori N, Falkow S (2002) Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417: 243–244. [DOI] [PubMed] [Google Scholar]
- 20. Joshua GW, Karlyshev AV, Smith MP, Isherwood KE, Titball RW, et al. (2003) A Caenorhabditis elegans model of Yersinia infection: biofilm formation on a biotic surface. Microbiology 149: 3221–3229. [DOI] [PubMed] [Google Scholar]
- 21. Matthysse AG, Stretton S, Dandie C, McClure NC, Goodman AE (1996) Construction of GFP vectors for use in Gram-negative bacteria other than Escherichia coli . FEMS Microbiol Lett 145: 87–94. [DOI] [PubMed] [Google Scholar]
- 22. Spangler C, Bohm A, Jenal U, Seifert R, Kaever V (2010) A liquid chromatography-coupled tandem mass spectrometry method for quantitation of cyclic di-guanosine monophosphate. J Microbiol Methods 81: 226–231. [DOI] [PubMed] [Google Scholar]
- 23. van Helden J (2003) Regulatory sequence analysis tools. Nucleic Acids Res 31: 3593–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Christensen GD, Simpson WA, Younger JJ, Baddour LM, Barrett FF, et al. (1985) Adherence of coagulase-negative Staphylococci to plastic tissue culture plates: a quantitative model for the adherence of Staphylococci to medical devices. J Clin Microbiol 22: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ali A, Rashid MH, Karaolis DK (2002) High-frequency rugose exopolysaccharide production by Vibrio cholerae . Appl Environ Microbiol 68: 5773–5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen Y, Dai J, Morris JG Jr, Johnson JA (2010) Genetic analysis of the capsule polysaccharide (K antigen) and exopolysaccharide genes in pandemic Vibrio parahaemolyticus O3:K6. BMC Microbiol 10: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, et al. (1999) Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis . Proc Natl Acad Sci U S A 96: 14043–14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Perry RD, Fetherston JD (1997) Yersinia pestis–etiologic agent of plague. Clin Microbiol Rev 10: 35–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Erickson DL, Jarrett CO, Wren BW, Hinnebusch BJ (2006) Serotype differences and lack of biofilm formation characterize Yersinia pseudotuberculosis infection of the Xenopsylla cheopis flea vector of Yersinia pestis . Journal of bacteriology 188: 1113–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hinnebusch BJ, Rudolph AE, Cherepanov P, Dixon JE, Schwan TG, et al. (2002) Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science 296: 733–735. [DOI] [PubMed] [Google Scholar]
- 31. Sun YC, Guo XP, Hinnebusch BJ, Darby C (2012) The Yersinia pestis Rcs phosphorelay inhibits biofilm formation by repressing transcription of the diguanylate cyclase gene hmsT . J Bacteriol 194: 2020–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chain PS, Carniel E, Larimer FW, Lamerdin J, Stoutland PO, et al. (2004) Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis . Proc Natl Acad Sci U S A 101: 13826–13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Erickson DL, Jarrett CO, Callison JA, Fischer ER, Hinnebusch BJ (2008) Loss of a biofilm-inhibiting glycosyl hydrolase during the emergence of Yersinia pestis . J Bacteriol 190: 8163–8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhou D, Yang R (2011) Formation and regulation of Yersinia biofilms. Protein Cell 2: 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative HPLC-MS/MS traces. c-di-GMP in a extract of WT (a) or Δfur (b), and a standard sample of c-di-GMP (c) in water at a concentration of 0.3 nM were detected by HPLC-MS/MS -di-GMP c-di-GMP. cXMP was used as the internal standard at a concentration of 50 ng/ml.
(JPG)