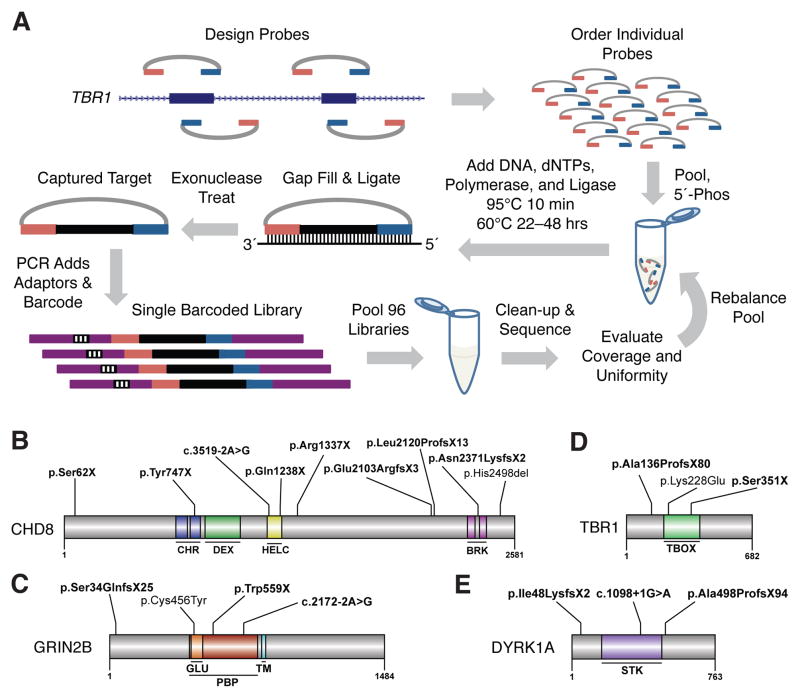
Fig. 1.
Massively multiplex targeted sequencing identifies recurrently mutated genes in ASD probands. (A) Schematic showing design and general workflow of a modified MIP method enabling ultra-low-cost candidate gene resequencing in very large cohorts (figs. S1–S7 and tables S1–S9) (10). (B to E) Protein diagrams of four genes with multiple de novo mutation events. Significant protein domains for the largest protein isoform are shown (colored regions) as defined by SMART (23) with mutation locations indicated. (B) CHD8. (C) GRIN2B. (D) TBR1. (E) DYRK1A. Bold variants are nonsense, frameshifting indels or at splice-sites (intron-exon junction is indicated). Domain abbreviations: CHR-chromatin organization modifier, DEX-DEAD-like helicases superfamily, HELC-helicase superfamily c-terminal, BRK-domain in transcription and CHROMO domain helicases, GLU-ligated ion channel L-glutamate- and glycine-binding site, PBP-eukaryotic homologs of bacterial periplasmic substrate binding proteins, TM-transmembrane, STK-serine-threonine kinase catalytic, TBOX-T-box DNA binding.