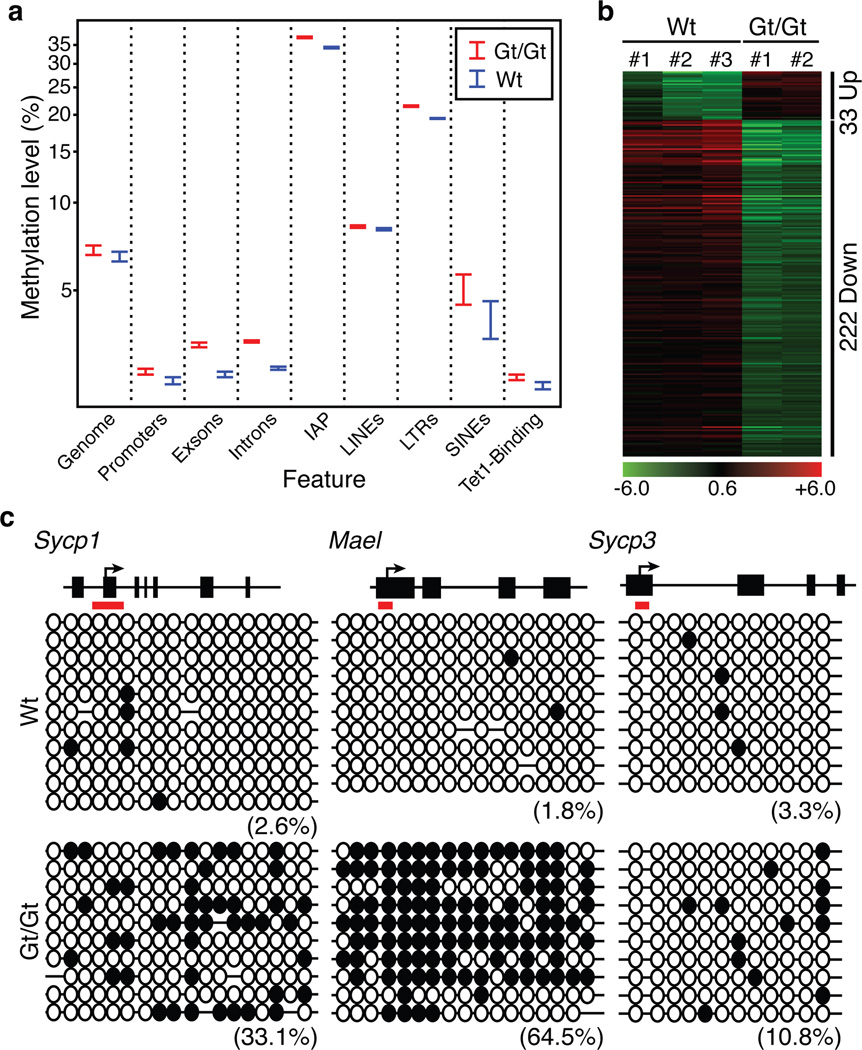
Figure 4. Whole genome bisulfite analysis of the effect of Tet1 knockout on DNA methylation in PGCs.
(a) Wt E13.5 female PGCs are globally hypomethylated and depletion of Tet1 only slightly increased the global DNA methylation level. Shown are the DNA methylation levels in the entire mouse genome as well as the various genomic regions. In the last column, the DNA methylation levels at the Tet1 bound regions identified in mES cells17 were compared.
(b) Heat map of the 255 differentially expressed and DMR associated genes.
(c) Bisulfite sequencing analysis of the Sycp1, Mael, and Sycp3 gene promoters of Tet1 binding site in wild-type and Tet1Gt/Gt PGCs. Each CpG is represented by a circle. Open and filled circles represent unmethylated or methylated, respectively. Percentages of DNA methylation are indicated.