Fig. 1.
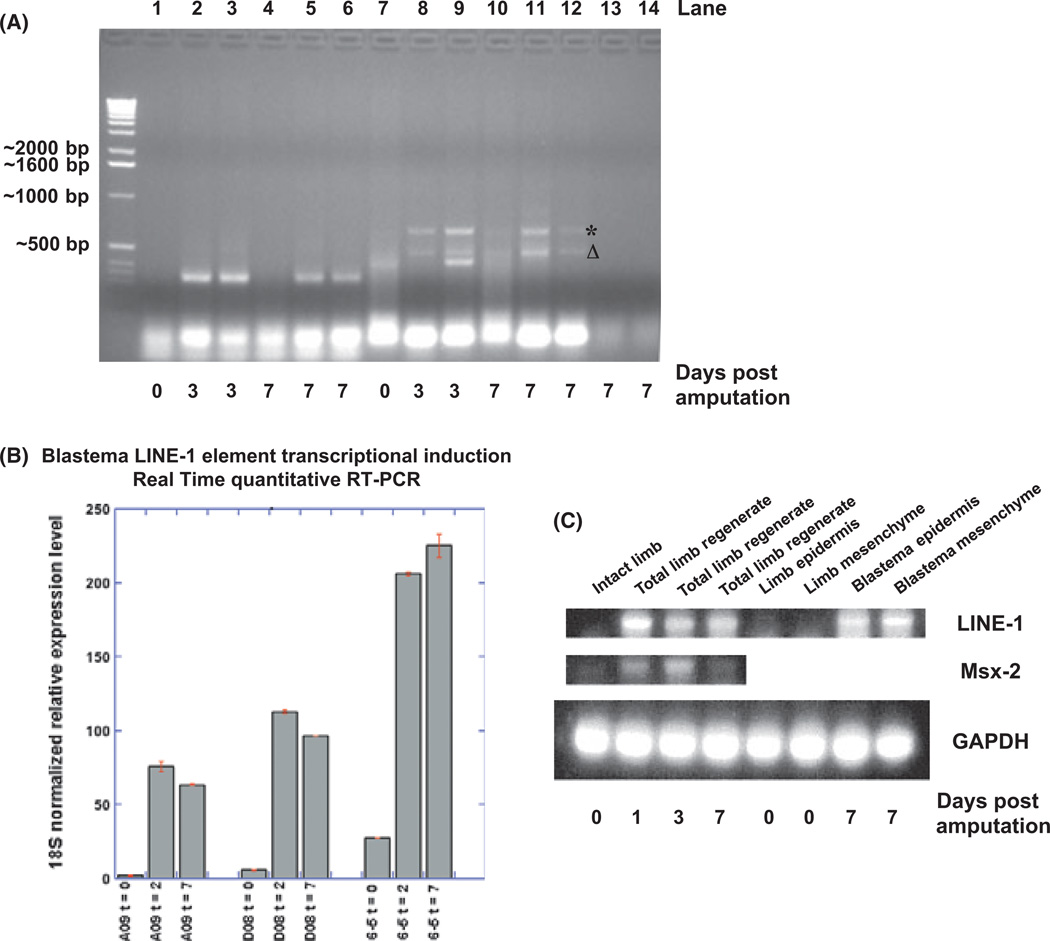
(A) Reverse transcription–polymerase chain reaction (RT–PCR) analysis primed by the DNA primers designed specifically for axolotl Oct4. Lanes 1–6 and 13 show the RT–PCR products resulting from the aOct4 primer set: aOct4 (81-106) and aOct4 (528-503). The nonspecific PCR products in these lanes were not characterized further. Lanes 7–13 were the RT–PCR products from another aOct4 primer set: aOct4 (741-768) and aOct4 (1068-1095). Sequencing results of the nonspecific RT–PCR products primed by aOct4 (741-768) and aOct4 (1068-1095) oligonucleotides revealed that they were either partial LINE-1 ORF1 (*) or ORF2 (Δ) cDNA fragments when the templates were derived from limb regenerates of different animals on day 3 or 7 days post-amputation (pa). Lanes 13 and 14 are the controls without primers. (B) Axolotl LINE-1 cDNA Real Time quantitative (q)PCR to examine LINE-1 transcriptional activity after limb amputation. A09 and D08 are the two sets of qPCR primers for LINE-1 ORF1, while 6–5 is a pair of primers for LINE-1 ORF2. t, days after limb amputation. (C) LINE-1 RT–PCR analysis on day 0, 3 or 7 pa. RT–PCR was primed by LINE-1-specific primers (P25R and P25F/26), LINE-1 transcriptional activity was examined in the regenerating epidermal and mesenchymal tissues (lanes 4–8), respectively, in addition to the total regenerating limb tissues (lanes 1–4). The transcriptional upregulation of another limb amputation-induced gene, Msx2 RNA was also examined by RT–PCR in parallel. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) RT–PCR served as loading controls.