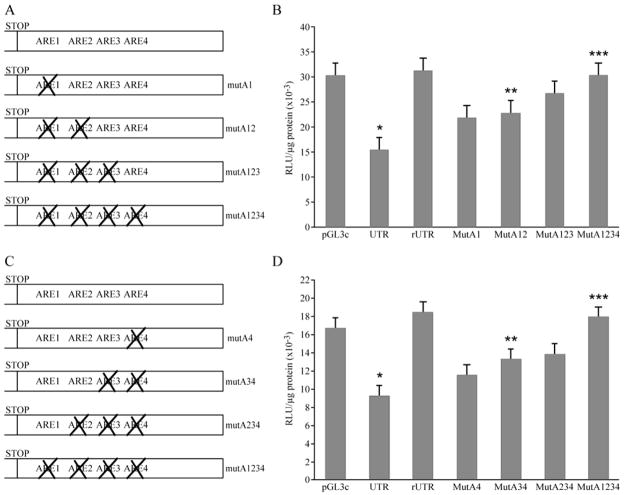
Fig. 4.
Mapping of the ARE elements responsible for the regulation of PTHrP 1-141 mRNA stability. A. Diagram of mutations introduced into the 3′-UTR to sequentially disrupt AREs in a 5′ to 3′direction. B. Transient transfections of HARA cells with mutated 3′-UTR-luciferase reporter constructs. HARA cells were transfected with 2 μg of either empty pGL3control vector or pGL3vector containing either the wild-type 3′-UTR in direct (pGL3c-UTR) or reverse (pGL3c-rUTR) orientations or the 3′-UTR with mutated AREs (pGL3c-MutA1, pGL3c-MutA12, pGL3c-MutA123 and pGL3c-MutA1234). After adjusting for multiple comparisons, UTR was significantly lower than GL3 (*, p-value < 0.0004), mutA12 was significantly greater than UTR (**, p-value = 0.009), mutA1234 was significantly greater than mutA12 (***, p-value = 0.0123) and mutA1234 was not significantly different from GL3 (p-value = 0.9958). C. Diagram of mutations introduced into 3′-UTR of 1-141 isoform to sequentially disrupt AREs in a 3′ to 5′ direction. D. Transient transfections of HARA cells with mutated 3′-UTR-luciferase reporter constructs. HARA cells were transfected with 2 μg of either empty pGL3control vector or pGL3vector containing either the wild-type 3′-UTR in direct (pGL3c-UTR) or reverse (pGL3c-rUTR) orientations or the 3′-UTR with mutated AREs (pGL3c-MutA4, pGL3c-MutA34, pGL3c-MutA234 and pGL3c-MutA1234). After adjusting for multiple comparisons, UTR was significantly lower than GL3 (*, p-value < 0.0004), mutA34 was significantly greater than UTR (**, p-value = 0.002), mutA1234 was significantly greater than mutA34 (***, p-value = 0.0006) and mutA1234 was not significantly different from GL3 (p-value = 0.29).