Figure 4.
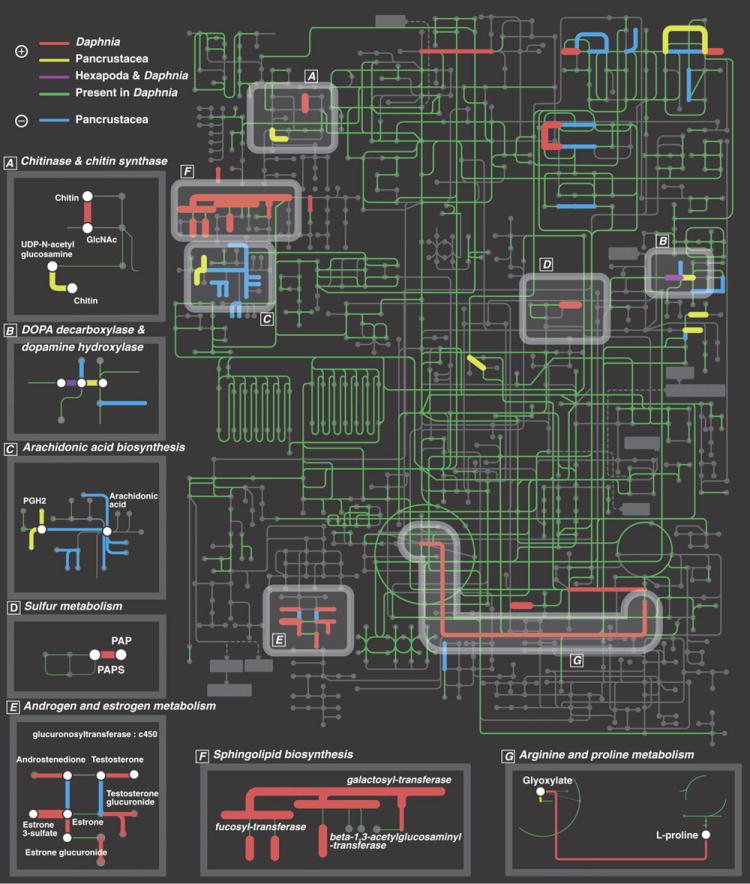
Map of global KEGG metabolic pathway in D. pulex showing significantly expanded or contracted gene families in metabolic pathways. Nodes and edges represent compounds and enzymes respectively. Expanded gene families in D. pulex (red); expanded gene families in Pancrustacea (yellow); independently expanded gene families in D. pulex and in insects (purple); contracted gene families in Pancrustacea (blue); and genes present in D. pulex (green). Amplification of gene families encoding each highlighted enzyme is supported by the Fisher exact test (thick edges are supported by Bonferroni correction), on the basis of the distribution of the number of genes encoding corresponding enzymes among Homo sapiens, Mus musculus, Gallus gallus and Tetraodon nigroviridis, Drosophila melanogaster, Apis mellifera and Anopheles gambiae. Emphasized pathways (A-G) include at least two cases of expanded interacting enzymes. The non-random co-expansion of interacting enzymes is supported by exact binomial test (p < 0.03) and by the node permutation test on 1,000 randomized metabolic networks (p<0.03).
