Fig. 4.
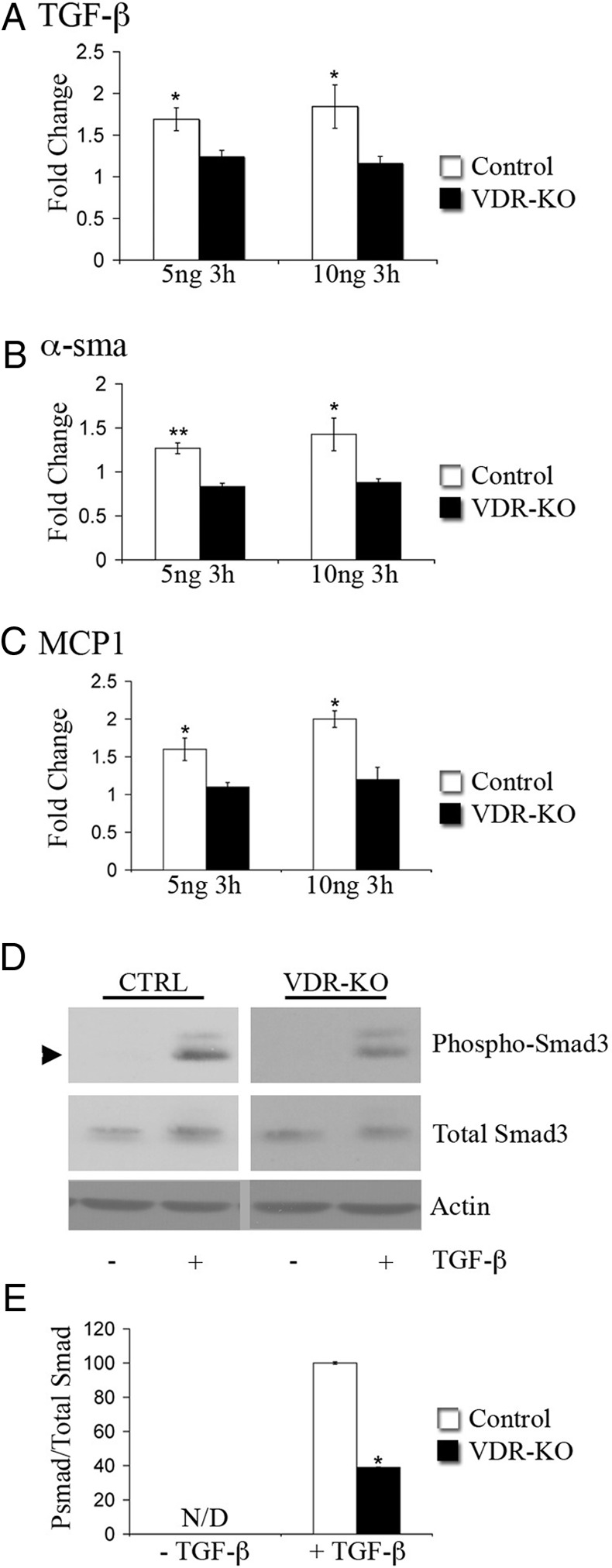
The VDR is required for the activation of TGF-β target gene expression in dermal fibroblasts. A–C, RNA isolated from primary wild-type (Control, white bars) or vdr−/− dermal fibroblasts(VDR-KO, black bars) treated for 3 h with 5 or 10 ng/ml TGF-β was subjected to qRT-PCR analysis for the genes indicated. Values are expressed as the relative fold change in expression of each transcript compared with unstimulated controls. RNA levels encoding each gene of interest were normalized for actin RNA in the same sample. Data shown are based on at least three independent RNA isolations ±sem. *, P ≤ 0.05, **, P ≤ 0.005. D, Total protein (7 μg) isolated from control (CTRL) or vdr−/− (VDR-KO) primary dermal fibroblasts treated with (+) or without (−) 10 ng TGF-β for 30 min was subjected to SDS-PAGE and immunoblotted for pSmad3, total Smad3, and actin. E, The ratio of pSmad3 to total Smad3 was determined by the quantitation of signal intensity of the relevant bands. The ratio of pSmad3 to total Smad3 of the TGF-β-treated vdr−/− dermal fibroblasts was normalized to that of the TGF-β-treated wild-type fibroblasts. Statistical significance was determined using the Student's t test, *, P ≤ 0.05. N/D, pSmad3 band was not detected in the absence of TGF-β. Data are representative of protein lysates obtained from four independent experiments.
