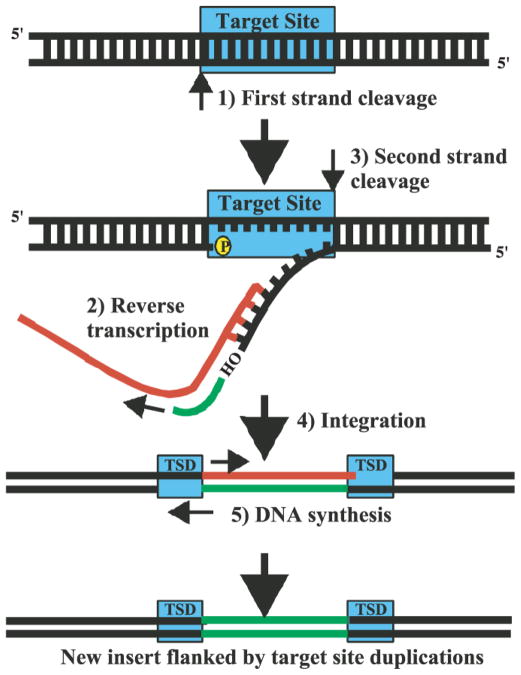
Figure I.
The retroposition mechanism. The sequence is initiated with a nicking of the first (bottom) DNA strand by an endonuclease (step 1). The new 3′ end serves as a primer for reverse transcription of a template RNA (red, step 2) [84]. Second strand cleavage (step 3) occurs during or after reverse transcription. After integration (step 4), the hybrid RNA is degraded, and second strand synthesis ensues (step 5). Newly synthesized DNA strands are in green. Target site duplications (TSDs) are hallmarks of retroposons. They occur because target site cleavage produces staggered ends, and the resulting small gaps are filled in on both sides of the insertion, giving rise to short flanking duplications. Adapted from Kazazian et al. [84], with permission.