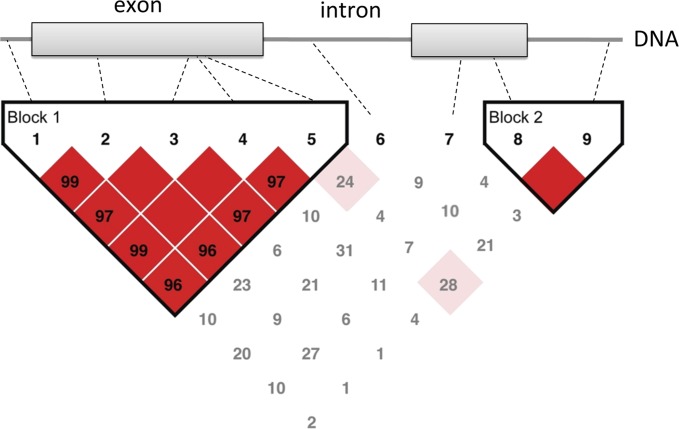
Figure 1.
Schematic illustrating the relationship of rare and common variants to linkage disequilibrium (LD) blocks in the genome. Linkage disequilibrium refers to the nonrandom associations of alleles at different loci. Variants 1–5, 8, and 9 are single-nucleotide polymorphisms (SNPs; >5% population frequency) whereas variants 6 and 7 are rare variants (<1% population frequency). SNPs 1–5 in block 1 are not independent and therefore in LD with each other, whereas they are not in LD with SNPs in block 2 (SNPs 8 and 9). Only one of the five SNPs in block 1 need be genotyped to capture the variation in block 1. Similarly, only one of two SNPs in block 2 must be genotyped to capture the variation in block 2. In addition to occurring infrequently, rare variants are not in LD with any common polymorphisms and therefore must be sequenced. Values in the LD map represent pair-wise 100 × D′ values of linkage disequilibrium. The blank squares represent D′ values of 1.0 (complete LD). Strong LD is indicated by red squares, whereas pink squares and white squares indicate uninformative and low confidence values, respectively.