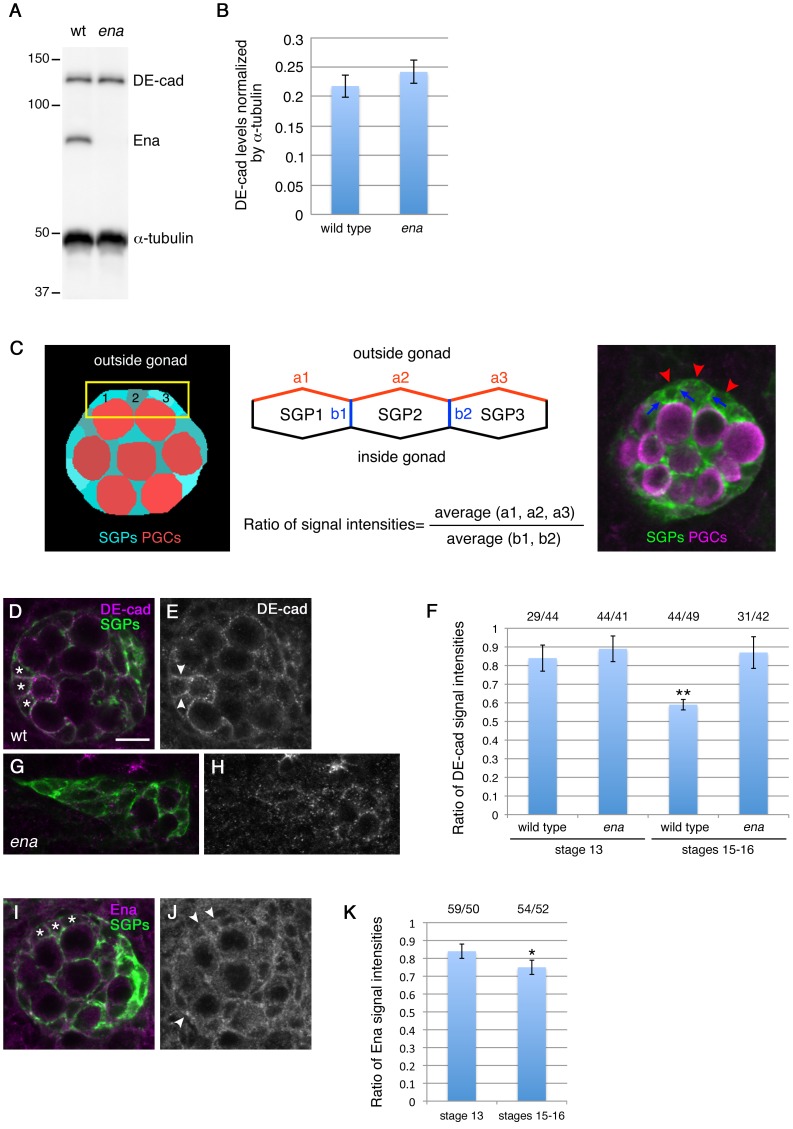
Figure 9. The ena mutation alters intracellular localization of DE-cad in SGPs.
(A, B) The DE-cad protein in control and ena mutant embryos was detected by Western Blot (A) and the expression level of DE-cad normalized against α-tubulin expression was plotted in the graph (B). (C–H) Wild-type and ena mutant SGPs were stained with the DE-cad antibody, and the signal intensity on the outer membrane (red) or SGP-SGP boundary (blue) was measured (middle and left panels). The ratio of the signal intensity on the outer membrane compared to that on the SGP-SGP boundary was calculated at stage 13 and stages 15–16 (middle panel). In wild type, DE-cad becomes enriched at the SGP-SGP boundary as gonad compaction proceeds (**P<0.01) (D–F). This process was affected in ena mutants (F–H). (I–K) Wild-type SGPs were stained with the anti-Ena antibody (I, J), and the ratio of the signal intensity on the outer membrane compared to that on the SGP-SGP boundary was plotted at stage 13 and stages 15–16 (K). Ena localization shifts towards the SGP-SGP boundary at stages 15–16 (*P<0.05). The number of outer membranes versus SGP-SGP boundaries scored is indicated above the graph (F, K). Error bars in (B, F, and K) represent standard error.