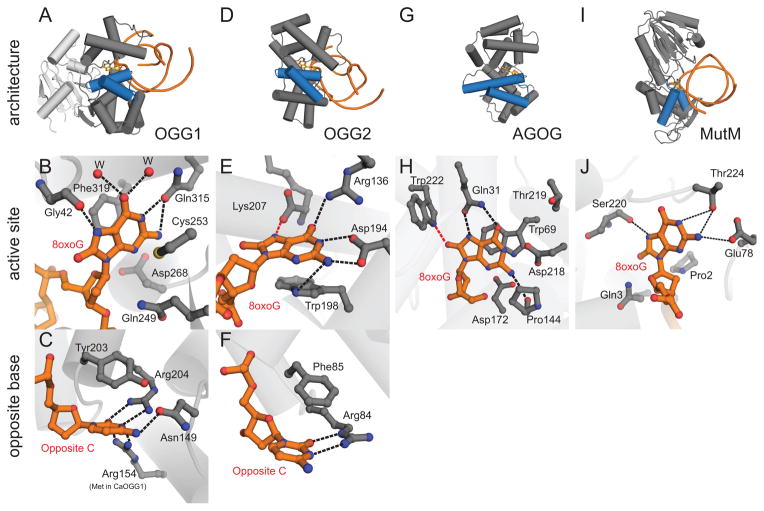
Figure 4.
Oxidative DNA glycosylases. (A–C) OGG1, represented by human OGG1 (PDB ID 1EBM), (D–F) OGG2, represented by MjOGG (PDB ID 3KNT), (G–H) Pyrobaculum aerophilum AGOG (PDB ID 1XQP) and (I–J) Geobacillus stearothermophilus MutM. The overall folds of each enzyme are shown on the top row (blue HhH motif), active sites on the second row, and opposing base on the bottom row. In the close-up views, the protein side-chains are grey and the DNA orange. Water molecules are represented by red spheres and hydrogen bonds are shown as dashed lines. (B) The human OGG1 8oxoG recognition pocket. The only 8oxoG specific contact is the hydrogen bond from the carbonyl group of Gly42 to the protonated N7 of 8oxoG. (C) The high specificity of hOGG1 for 8oxoG•C base pairs can be rationalized by the 5 hydrogen bonds between the opposite cytosine and 3 side chains. (E) In MjOGG, the 8oxoG N7 donates a hydrogen bond (red dashed line) to the C-terminal Lys207 carboxylate. (F) The opposite cytosine in MjOGG is contacted by only one side chain. (H) 8oxoG nucleoside bound inside the AGOG active site, with a unique 8oxoG-specific contact to Trp222 (red dashed line). (J) Active site of MutM (PDB ID 1R2Y) shows multiple contacts to 8oxoG but lacks the aromatic residues seen in the OGG1, OGG2, and AGOG enzymes.