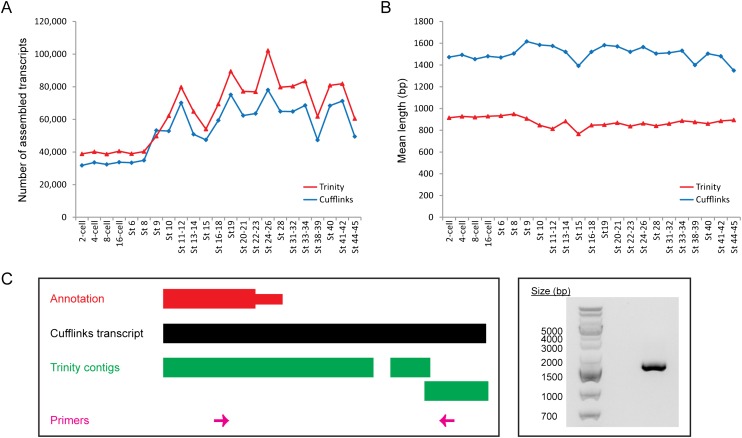
Figure 7.
Ab initio transcript assembly with a reference genome (Cufflinks) results in fewer but longer contigs than de novo transcript assembly without a reference genome (Trinity). (A) Number of contigs assembled by Cufflinks (blue diamonds) and Trinity (red triangles) at every developmental stage. Trinity almost always reconstructs a larger number of transcripts than Cufflinks. (B) The mean length of Cufflinks transcripts (blue diamonds) and Trinity contigs (red triangles) at each stage. On average, a transcript reconstructed by Cufflinks is 628 bp longer than a contig assembled by Trinity. (C) The Cufflinks transcript for the ccnK gene is more accurate than the corresponding Trinity contig. (Left) A schematic of the genomic locus of ccnK at the 3′ end. (Right) A gel image showing the result of a PCR performed with the primers depicted in pink at the left. The PCR product shown was further sequenced to confirm that it was indeed the intended target.