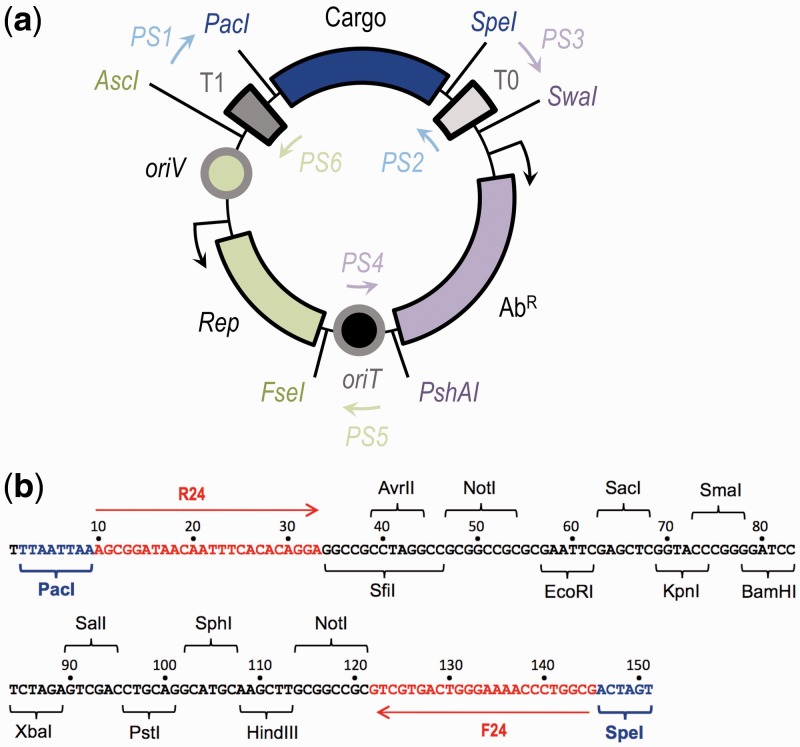
Figure 2.
Overall organization of structure of SEVA plasmids. (a) SEVA vectors are formed by three variable modules: a cargo (blue), a replication origin (green) and an antibiotic marker (magenta). Enzymes used to change the functional DNA segments are shown in the same color code and modules are separated by three permanent regions, which are shared by all vectors, the T0 and T1 transcriptional terminators and the oriT conjugation origin. All universal primers (PS1–PS6) are placed within the invariable backbone and are used to sequence/check the variable modules. (b) The structure of the default SEVA cargo. Cargos are cloned as PacI/SpeI fragments. The default segment contains the typical pUC18 polylinker enzymes from EcoRI to HindIII (the completely ordered restriction enzyme list is highlighted in the figure). Additional enzymes (i.e. SfiI, AvrII and NotI) are placed outside of the polylinker for specific cloning purposes. Within the cargo sequence, the enzyme recognition sites and the hybridization site of the universal M13 (R24/F24) primers are shown.