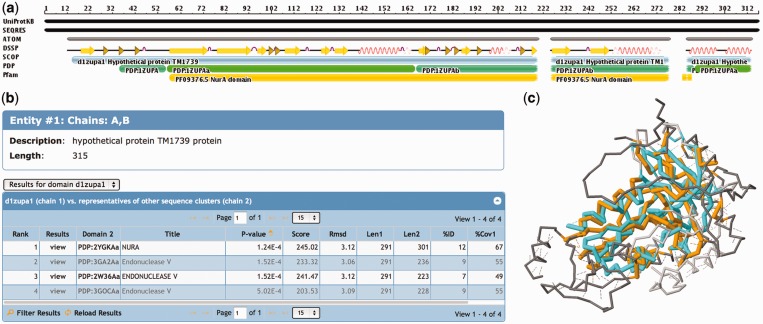
Figure 2.
Domain-based structural alignment database search results for PDB ID 1ZUP (30), a hypothetical protein and a putative nuclease. (a) The sequence is shown with different annotations, starting with the UniProtKB sequence at the top. Most of this sequence has been resolved (SEQRES) with ATOM records available in the PDB, with the exception of a region at the N-terminus and two short regions toward the C-terminus of the protein. SCOP provides an annotation of a single domain protein, the PDP software assigns two domains to this protein and Pfam detects a NurA domain motif. (b) The hits listed in the table show similarities to a NurA structure, as well as several endonucleases. (c) When viewing the details of the first alignment [1ZUP shown in orange and NurA domain of 2YGK (31) in cyan], one notices that several loop and helical regions are not conserved between the two structures, but the core is well conserved. Structural alignments are accessible from the 3D Similarity tab of any entry’s Structure Summary page.