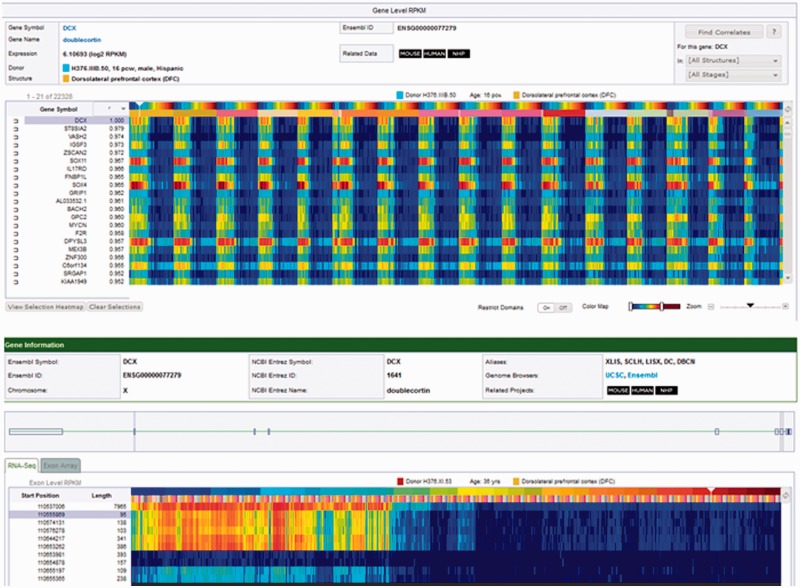
Figure 1.
Transcriptome RNA sequencing heat maps. The transcriptome heat map is a visualization of the expression values for the returned gene of interest (top panel). In this example, the Find Correlates search returns (search by example) are shown for DCX (doublecortin), which has high expression during prenatal development. The heat map data is presented as a matrix with brain structure (by developmental stage) on the x-axis and genes on the y-axis. Brain structures are organized in ontological order. Clicking on the toggle button will change the initial sorting parameter from structure to developmental stage. In this example, data is sorted by structure, and then by the youngest to oldest developmental stage within each structure. Each column of the heat map represents a tissue sample. The colors of the heat map are expression values, transformed to a log2 scale. The color scale ranges from dark blue, representing low expression, and passes through cyan, yellow and orange and finally to dark red, representing high expression. The lower panel presents the exon heat map view of the RNA sequencing data for DCX. Each row of the heat map in the RNA sequencing data represents an exon. Exons in the RNA sequencing data are labeled by the chromosome start position and exon length. Positioning the mouse over an exon highlights the corresponding exon in the schematic composite gene model.