Figure 1.
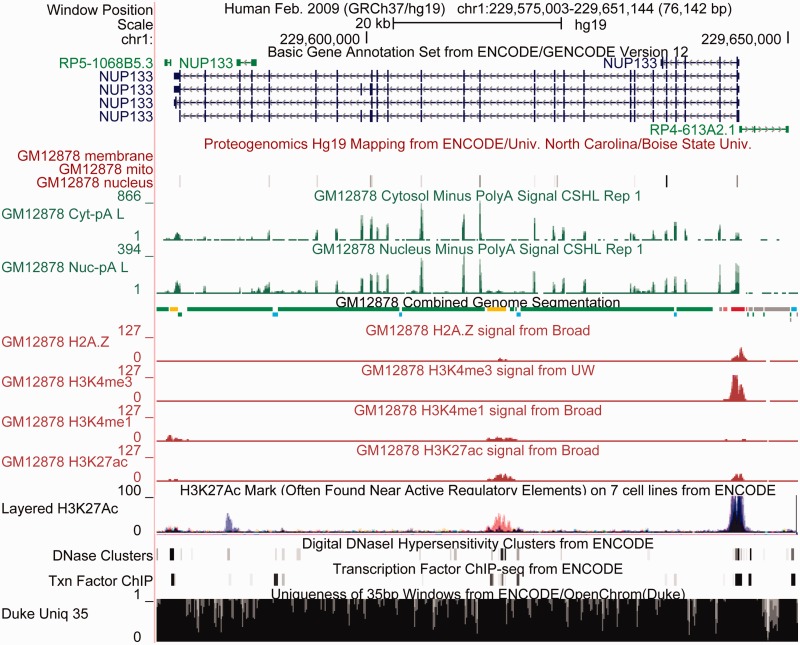
ENCODE data displayed in the UCSC Genome Browser together with annotations from the ENCODE Analysis Hub in the region of the nucleoporin gene NUP133 demonstrate the power this diversity of data provides for visual interpretation. The GENCODE Basic gene set shows this gene having four protein-coding splice variants and three smaller non-coding transcripts nearby. The proteogenomics track shows support for many of the coding exons, with protein localized in the nucleus, but not in plasma membrane or mitochondria. The long polyA RNA signal shows strong peaks over the exons and low intron signal in the cytosol, with greater signal in the nucleus. This is expected because nuclear mRNAs are not all completely spliced. The Combined Genome Segmentation integrates signal from many histones and classifies regions into those with characteristics of promoters (red), enhancers (yellow), insulators (blue), transcribed regions (green) and repressed (gray). Below are signal tracks from four of the eight histone modifications used as input to the segmentation. The promoter and transcribed regions agree with the RNA evidence, and like the RNA evidence show no evidence of transcription of the non-coding gene to the right of NUP133. Underneath the GM12878 histone signals is a track that overlays one of the histone signals, H3K27Ac, in seven different cell lines (with GM12878 shown in red). A peak in H3K27Ac appears at the enhancer, but as is often the case with enhancers, this appears to be relatively cell specific in contrast to the larger peak near the promoter, where the black coloration indicates the peak is shared by many cell types. The DNAse hypersensitivity and transcription factor tracks also provide evidence for both promoter and enhancer. Finally the mappability track indicates regions where short reads are not uniquely mappable, indicating the data are incomplete and therefore harder to interpret. Although most of this region is mappable, there are many small regions throughout and one larger region on the right where mapping is problematic. Overall, the ENCODE data in this region show strong evidence that this is a nuclear-localized protein-coding gene with a promoter that is used in a wide variety of cell types, and is likely to be regulated by tissue-specific enhancers as well.