Abstract
We here present The Online Protein Processing Resource (TOPPR; http://iomics.ugent.be/toppr/), an online database that contains thousands of published proteolytically processed sites in human and mouse proteins. These cleavage events were identified with COmbinded FRActional DIagonal Chromatography proteomics technologies, and the resulting database is provided with full data provenance. Indeed, TOPPR provides an interactive visual display of the actual fragmentation mass spectrum that led to each identification of a reported processed site, complete with fragment ion annotations and search engine scores. Apart from warehousing and disseminating these data in an intuitive manner, TOPPR also provides an online analysis platform, including methods to analyze protease specificity and substrate-centric analyses. Concretely, TOPPR supports three ways to retrieve data: (i) the retrieval of all substrates for one or more cellular stimuli or assays; (ii) a substrate search by UniProtKB/Swiss-Prot accession number, entry name or description; and (iii) a motif search that retrieves substrates matching a user-defined protease specificity profile. The analysis of the substrates is supported through the presence of a variety of annotations, including predicted secondary structure, known domains and experimentally obtained 3D structure where available. Across substrates, substrate orthologs and conserved sequence stretches can also be shown, with iceLogo visualization provided for the latter.
More than two percent of all human and mouse genes encode proteases. These enzymes control many biological processes and are of crucial importance for relatively simple processes such as food digestion, as well as for highly regulated proteolytic cascades such as controlled cell death or blood coagulation. In addition, misregulated protease activities add to the severity of several pathologies, including cancer, cardiovascular and inflammatory diseases. It is commonly recognized that a more detailed understanding of protease-controlled or protease-affected processes can be achieved by extending our overall knowledge on proteases, their (preferred) substrates and specificities (1).
The N-terminal COFRADIC (COmbinded FRActional DIagonal Chromatography) technique developed in our laboratory enables isolation and identification of protein N-terminal peptides using peptide chromatography and mass spectrometry (MS) (2). Given that protein processing induces new N- and C-terminal protein ends, the resulting neo-N-terminal peptides are also isolated and identified, and represent proxies for the actual cleavage position in the protease substrate (3). Recently, a similar C-terminal COFRADIC technique was developed to select and identify (neo-) C-terminal peptides, thus also identifying processing events (4). These COFRADIC techniques made the identifications of large numbers of processing events possible, and these techniques are applicable for both individual proteases and in cellular setups in which several proteases are active (3,5–12). It should be noted that besides the COFRADIC technologies, other mass spectrometry-based technologies were recently introduced to identify protein processing events [recently reviewed in (13)].
A number of databases are currently available that disseminate protein processing events. The MEROPS database is specialized in proteases and their classification, and stores substrates linked to proteases (14). However, this database does not easily allow a user to perform specific meta-analyses such as a direct comparison among substrates. PMAP/CutDB on the other hand is a community-driven (Wikipedia style) database, implying that any scientist can add new substrates or substrate predictions (15,16). The intrinsic disadvantage is that the quality of the reported substrates cannot be guaranteed, especially because the original data leading to the discovery of a substrate can only be accessed by using the provided links to the original article. CASBAH, developed in 2007 as part of a review article on proteolytic processes in dying cells, stores processing sites of caspases only (17). It is also important to note that a processing event in a substrate stored in CutDB or CASBAH is not necessarily linked to the actual cleavage position in the substrate. Recently, the TopFIND 2.0 database has been released as well, offering a protein-centric knowledgebase on protein termini, including modifications and processing events (18). TopFIND comes equipped with data mining and analysis tools, but is ultimately based on curated (imported) data from experimental data sets and existing databases, losing any direct connection to the underlying experimental data in the process. As such, it resembles the UniProtKB/Swiss-Prot model, albeit with more integrated analysis tools. Finally, the ApoptoProteomics database was also recently launched, but as can be derived from its name, this database centralizes on protein processing found in apoptotic cells from different origins (19).
In conclusion, no single database exists today that provides complete data provenance, an essential feature in guaranteeing data quality, with only TopFIND 2.0 and ApoptoProteomics providing support for further (meta-) analyses on both proteases and their substrates.
We here present The Online Protein Processing Resource (TOPPR) that stores published processing events identified in our laboratory by N- and C-terminal COFRADIC technologies. TOPPR makes our data available through an easy and intuitive analysis platform. Furthermore, the application provides a user interface that is specifically tailored to verify the actual MS/MS data that led to the identification of the reported processed sites. In fact, the Mascot identification score (20), corresponding threshold score and confidence level are easily checked for every peptide reporting a processing event. Additionally, the b- and y-ion annotated MS/MS spectrum can be viewed and downloaded using the PRIDE spectrum viewer (21). These annotations are derived from the underlying ms_lims processing pipeline (22) that is in turn built on the MascotDatfile library for reading and interpreting Mascot search results (23). Full data provenance is thus guaranteed, making it simple for the user to check data quality. Note that this implies that TOPPR is exclusively dedicated to displaying experimentally observed cleavages sites, and that the system does not include any predicted cleavage sites. The focus on empirical mass spectrometry-based proteomics data also means that TOPPR will under-represent any cleavages sites that are difficult to detect with this technology, notably in the case of heavily modified peptides. TOPPR also supports user-level security, allowing data to be kept private to one or more authenticated users before publication. All information in TOPPR is stored in a MySQL database (see Supplementary Figure S1 for the relational schema), and query results are generated by JavaServer Pages and Java Servlet technologies running on an Apache Tomcat server infrastructure. TOPPR is released under the permissive Apache2 open source license, and all source code can be downloaded from http://code.google.com/p/toppr/.
At the time of writing, TOPPR contains 2234 substrates, for 18 studied treatments or peptidases, resulting in 27 147 cleavages. To navigate these data, TOPPR provides three different search methods. The first method, the parameter search, is used to find all substrates for one treatment or a combination of treatments, where a treatment corresponds to either a cellular stimulus in an in vivo/in cellulo assay or, alternatively, a protease used in an in vitro assay (i.e. a protease added to a cell lysate). The user selects one or more treatments from a list of published treatments, and can perform a range of set operations on these to create specific queries. Furthermore, this query interface allows even more fine-grained retrieval options to be specified, including combinations of treatments that result in the same site being cleaved (processing ‘hot spots’). Second, a UniProtKB/Swiss-Prot search is provided by which users can use a UniProtKB/Swiss-Prot accession number, a UniProtKB/Swiss-Prot entry name or a fraction of the corresponding protein description as the search string. This method will reveal all stored processed sites linked to the specified substrate. Third, a motif search enables users to search for substrates containing processed sites that match the user-defined protease specificity profile. This specificity profile is defined in two parts: a pre-site (non-primed sites) and a post-site motif (primed sites) (24), with each motif defined using a simplified regular expression syntax.
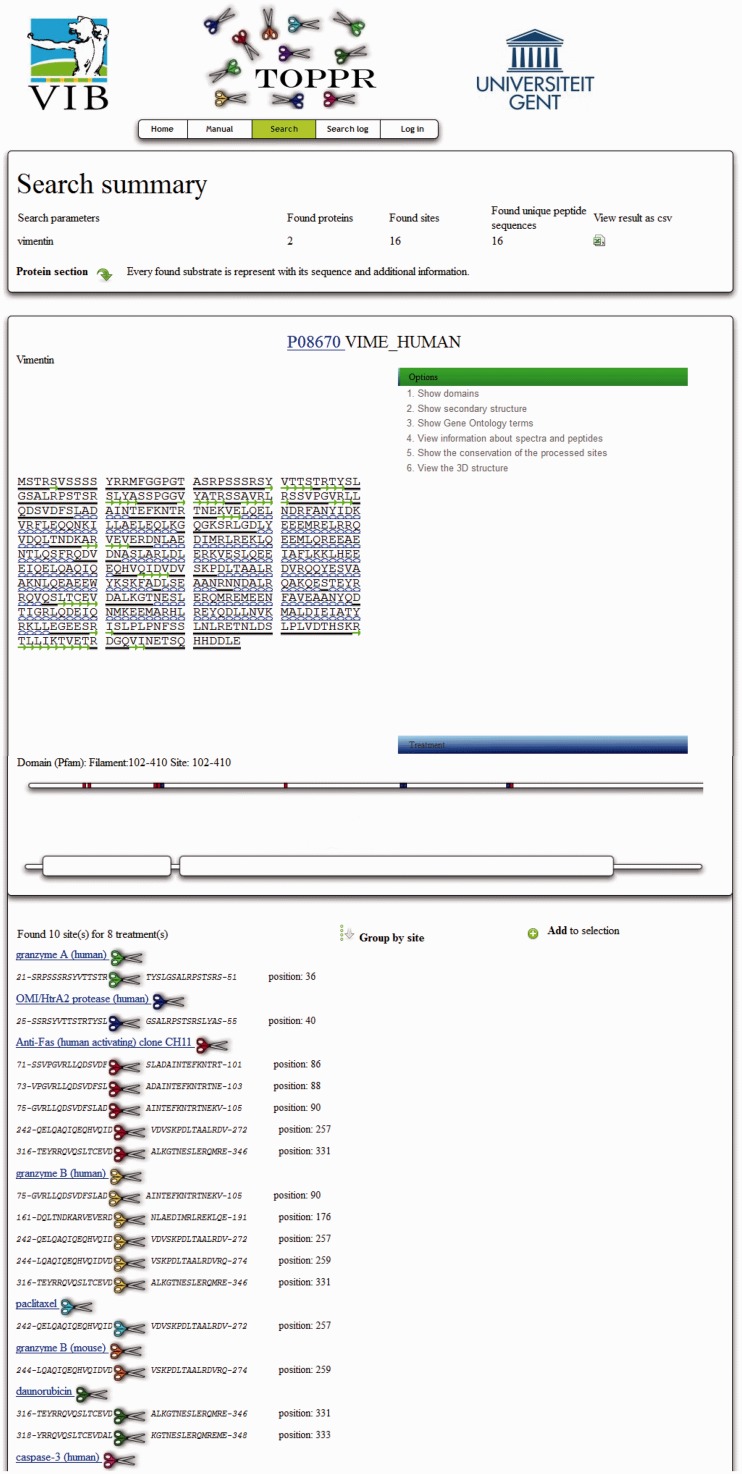
TOPPR also supports two types of analysis: analysis of the processed sites and corresponding protease specificity and, in addition, detailed analysis of individual substrates. The analysis of processed sites to infer protease specificity can be carried out by using integrated tools like iceLogo [probability-based visualization of significantly enriched/depleted residues in aligned sequences (25)], Weblogo [sequence logos (26)], PoPS [prediction of protease specificity (27)] and JalView [multiple sequence viewer and analysis tool (28)]. The list of processed sites used as input for these tools is extracted from TOPPR through the data retrieval options listed earlier, or as a manually selected subset of sites. The detailed analysis of individual substrates, on the other hand, is provided in TOPPR through a variety of integrated substrate metadata whenever available. First of all, processing events by different proteases are readily visualized using the substrate sequence view of TOPPR (see Figure 1). Additionally, Smart (29) and Pfam (30) annotations can be shown alongside reported processing events at the substrate level, thus indicating possible processing-derived interference with substrate function. Conservation of processing events among different species can be studied via a built-in function that globally aligns the surrounding sequences of processed sites in all known substrate homologs or orthologs (found in the HomoloGene database). Where available, TOPPR provides 3D structures (visualized with JMol) of the substrates. Processing events are indicated in these structures using a ball and stick configuration, whereas the rest of the polypeptide chain is in the cartoon configuration. This visualization makes it straightforward to assess the processing event in the context of the substrate’s 3D structure. Finally, the UniProtKB/Swiss-Prot annotated secondary structure elements, or, if unavailable, a secondary structure prediction (31) can be shown in the sequence view, facilitating substrate examination in the absence of 3D structures.
Figure 1.
The substrate sequence view in TOPPR. This display provides an overview of the protein sequence and links to dynamic annotations (here secondary structure and domain annotation have been selected). A protein bar representation, below the sequence, represents the full length of the protein with processing events indicated; immediately underneath, the domain visualization is shown. Below this protein-centric sequence view, details are shown on the individual peptides that were found to represent the annotated cleavage sites. Each peptide sequence in turn links to a peptide-centric view, where motif analyses, mass spectrometry data and all matching proteins can be found. Note that the peptide is annotated with its start and end coordinates on the protein.
The TOPPR database is continuously updated with novel findings, keeping track of all published protease cleavage sites identified by COFRADIC (or related) technologies in our laboratory. Over time, more treatments and substrates will therefore be included, leading to an increasingly comprehensive database of cleavage sites for the most abundant eukaryotic intracellular proteases (e.g. human and mouse caspases, granzymes, calpains and cathepsins). Furthermore, through the ability to transmit the data associated with an entire project from one ms_lims system to another via the Internet, TOPPR can easily receive incoming data from third parties. Users of ms_lims need only contact the authors for a username and password to connect to the TOPPR-linked ms_lims installation at the authors’ laboratories, at which point the project transmission application of ms_lims allows the data to be transmitted with a single click, ensuring its downstream uptake in TOPPR as well. Note that the reliance on ms_lims as the underlying data processing and management platform implicitly ensures consistency and comparable quality across all assembled data. Apart from its role as a data storage system, TOPPR also serves as a powerful exploration platform to verify data quality, assess protease specificity, perform processing site motif analyses and carry out detailed substrate analyses. An online user manual provides detailed information on the available search methods and types of analysis. TOPPR thus provides a powerful platform for discovery and analysis to both protease researchers and scientists studying a single substrate.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure 1.
FUNDING
Ghent University Multidisciplinary Research Partnership ‘Bioinformatics: from nucleotides to networks’; Fund for Scientific Research (FWO)—Flanders (Belgium) (postdoctoral research fellowship to F.I., P.V.D. and K.H.); Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen) (PhD to K.P.); ProteomeXchange project, funded by the European Union 7th Framework Program under grant agreement number [260558 to N.H.]; PRIME-XS project, funded by the European Union 7th Framework Program under grant agreement number [262067 to K.G. and L.M.]. Funding for open access charge: VIB, Ghent, Belgium.
Conflict of interest statement. None declared.
REFERENCES
- 1.Puente XS, Sánchez LM, Overall CM, López-Otín C. Human and mouse proteases: a comparative genomic approach. Nat. Rev. Genet. 2003;4:544–548. doi: 10.1038/nrg1111. [DOI] [PubMed] [Google Scholar]
- 2.Gevaert K, Goethals M, Martens L, Van Damme J, Staes A, Thomas G, Vandekerckhove J. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat. Biotechnol. 2003;21:566–569. doi: 10.1038/nbt810. [DOI] [PubMed] [Google Scholar]
- 3.Van Damme P, Martens L, Van Damme J, Hugelier K, Staes A, Vandekerckhove J, Gevaert K. Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat. Methods. 2005;2:771–777. doi: 10.1038/nmeth792. [DOI] [PubMed] [Google Scholar]
- 4.Van Damme P, Staes A, Bronsoms S, Helsens K, Colaert N, Timmerman E, Aviles FX, Vandekerckhove J, Gevaert K. Complementary positional proteomics for screening substrates of endo- and exoproteases. Nat. Methods. 2010;7:512–515. doi: 10.1038/nmeth.1469. [DOI] [PubMed] [Google Scholar]
- 5.Vande Walle L, Van Damme P, Lamkanfi M, Saelens X, Vandekerckhove J, Gevaert K, Vandenabeele P. Proteome-wide identification of HtrA2/Omi substrates. J. Proteome Res. 2007;6:1006–1015. doi: 10.1021/pr060510d. [DOI] [PubMed] [Google Scholar]
- 6.Impens F, Van Damme P, Demol H, Van Damme J, Vandekerckhove J, Gevaert K. Mechanistic insight into taxol-induced cell death. Oncogene. 2008;27:4580–4591. doi: 10.1038/onc.2008.96. [DOI] [PubMed] [Google Scholar]
- 7.Lamkanfi M, Kanneganti T-D, Van Damme P, Vanden Berghe T, Vanoverberghe I, Vandekerckhove J, Vandenabeele P, Gevaert K, Núñez G. Targeted peptidecentric proteomics reveals caspase-7 as a substrate of the caspase-1 inflammasomes. Mol. Cell. Proteomics. 2008;7:2350–2363. doi: 10.1074/mcp.M800132-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Damme P, Maurer-Stroh S, Plasman K, Van Durme J, Colaert N, Timmerman E, De Bock P-J, Goethals M, Rousseau F, Schymkowitz J, et al. Analysis of protein processing by N-terminal proteomics reveals novel species-specific substrate determinants of granzyme B orthologs. Mol. Cell. Proteomics. 2008;8:258–272. doi: 10.1074/mcp.M800060-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Demon D, Van Damme P, Vanden Berghe T, Deceuninck A, Van Durme J, Verspurten J, Helsens K, Impens F, Wejda M, Schymkowitz J, et al. Proteome-wide substrate analysis indicates substrate exclusion as a mechanism to generate caspase-7 versus caspase-3 specificity. Mol. Cell. Proteomics. 2009;8:2700–2714. doi: 10.1074/mcp.M900310-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Impens F, Colaert N, Helsens K, Ghesquiere B, Timmerman E, De Bock P-J, Chain BM, Vandekerckhove J, Gevaert K. A quantitative proteomics design for systematic identification of protease cleavage events. Mol. Cell. Proteomics. 2010;9:2327–2333. doi: 10.1074/mcp.M110.001271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Damme P, Maurer-Stroh S, Hao H, Colaert N, Timmerman E, Eisenhaber F, Vandekerckhove J, Gevaert K. The substrate specificity profile of human granzyme A. Biol. Chem. 2010;391:983–997. doi: 10.1515/BC.2010.096. [DOI] [PubMed] [Google Scholar]
- 12.Kaiserman D, Buckle AM, Van Damme P, Irving JA, Law RHP, Matthews AY, Bashtannyk-Puhalovich T, Langendorf C, Thompson P, Vandekerckhove J, et al. Structure of granzyme C reveals an unusual mechanism of protease autoinhibition. Proc. Natl Acad. Sci. USA. 2009;106:5587–5592. doi: 10.1073/pnas.0811968106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Impens F, Vandekerckhove J, Gevaert K. Who gets cut during cell death? Curr. Opin. Cell Biol. 2010;22:859–864. doi: 10.1016/j.ceb.2010.08.021. [DOI] [PubMed] [Google Scholar]
- 14.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Igarashi Y, Eroshkin A, Gramatikova S, Gramatikoff K, Zhang Y, Smith JW, Osterman AL, Godzik A. CutDB: a proteolytic event database. Nucleic Acids Res. 2007;35:D546–D549. doi: 10.1093/nar/gkl813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Igarashi Y, Heureux E, Doctor KS, Talwar P, Gramatikova S, Gramatikoff K, Zhang Y, Blinov M, Ibragimova SS, Boyd S, et al. PMAP: databases for analyzing proteolytic events and pathways. Nucleic Acids Res. 2009;37:D611–D618. doi: 10.1093/nar/gkn683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lüthi AU, Martin SJ. The CASBAH: a searchable database of caspase substrates. Cell Death Differ. 2007;14:641–650. doi: 10.1038/sj.cdd.4402103. [DOI] [PubMed] [Google Scholar]
- 18.Lange PF, Huesgen PF, Overall CM. TopFIND 2.0—linking protein termini with proteolytic processing and modifications altering protein function. Nucleic Acids Res. 2012;40:D351–D361. doi: 10.1093/nar/gkr1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arntzen MØ, Thiede B. ApoptoProteomics, an integrated database for analysis of proteomics data obtained from apoptotic cells. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.010447. M111.010447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 21.Vizcaíno JA, Côté R, Reisinger F, Foster JM, Mueller M, Rameseder J, Hermjakob H, Martens L. A guide to the Proteomics Identifications Database proteomics data repository. Proteomics. 2009;9:4276–4283. doi: 10.1002/pmic.200900402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helsens K, Colaert N, Barsnes H, Muth T, Flikka K, Staes A, Timmerman E, Wortelkamp S, Sickmann A, Vandekerckhove J, et al. ms_lims, a simple yet powerful open source laboratory information management system for MS-driven proteomics. Proteomics. 2010;10:1261–1264. doi: 10.1002/pmic.200900409. [DOI] [PubMed] [Google Scholar]
- 23.Helsens K, Martens L, Vandekerckhove J, Gevaert K. MascotDatfile: an open-source library to fully parse and analyse MASCOT MS/MS search results. Proteomics. 2007;7:364–366. doi: 10.1002/pmic.200600682. [DOI] [PubMed] [Google Scholar]
- 24.Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 25.Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. Improved visualization of protein consensus sequences by iceLogo. Nat. Methods. 2009;6:786–787. doi: 10.1038/nmeth1109-786. [DOI] [PubMed] [Google Scholar]
- 26.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyd S, Pike R, Rudy G, Whisstock J, Garcia De La Banda M. PoPS: a computational tool for modeling and predicting protease specificity. J. Bioinform. Comput. Biol. 2005;3:551–585. doi: 10.1142/s021972000500117x. [DOI] [PubMed] [Google Scholar]
- 28.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandonia J-M. StrBioLib: a Java library for development of custom computational structural biology applications. Bioinformatics. 2007;23:2018–2020. doi: 10.1093/bioinformatics/btm269. [DOI] [PMC free article] [PubMed] [Google Scholar]