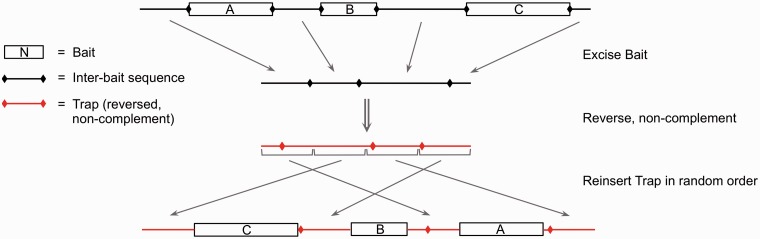
Figure 1.
Construction of the overextension trap. Bait sequences (a conservative set of bases matched by nhmmer + Dfam and both cross_match and rmblastn with consensus sequences) were placed in inverted order. Inter-bait sequences were concatenated into a long stretch of sequence that was reversed without complementation and divided into equal sized blocks, which were then placed in random order between the bait sequences.