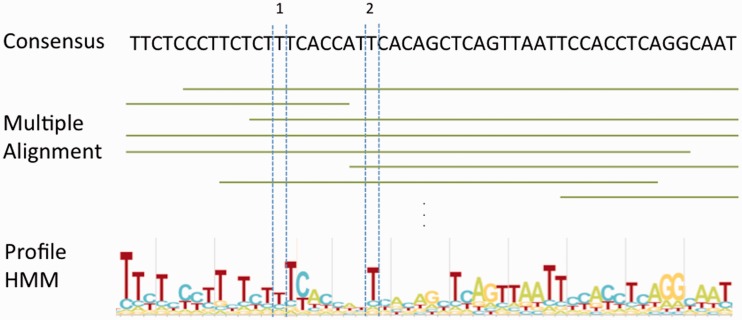
Figure 2.
Schematic representation of the creation of the multiple sequence alignment and profile HMM for a Dfam entry. The consensus and HMM logo correspond to positions 253–304 of Tigger16a (DF0000028), and highlight the difference between the abilities of HMM and consensus to represent positional residue conservation — a consensus treats all majority rule decisions as equivalent, while a profile HMM enables position-specific scoring based on conservation. In this case, the position labelled with (1) has a slight preference for ‘T’, but will not substantially reward a ‘T’ or penalize any other nucleotide; meanwhile the position labelled with (2) shows a strong preference for ‘T’, and will provide high reward for a matching ‘T’, and a strong penalty for any other nucleotide.